Heterocyclic modulators of lipid synthesis
A fatty acid synthase and cycloalkyl technology, which can be used in peptide/protein components, active ingredients of heterocyclic compounds, and medical preparations containing active ingredients, etc., and can solve the problem of not being approved.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
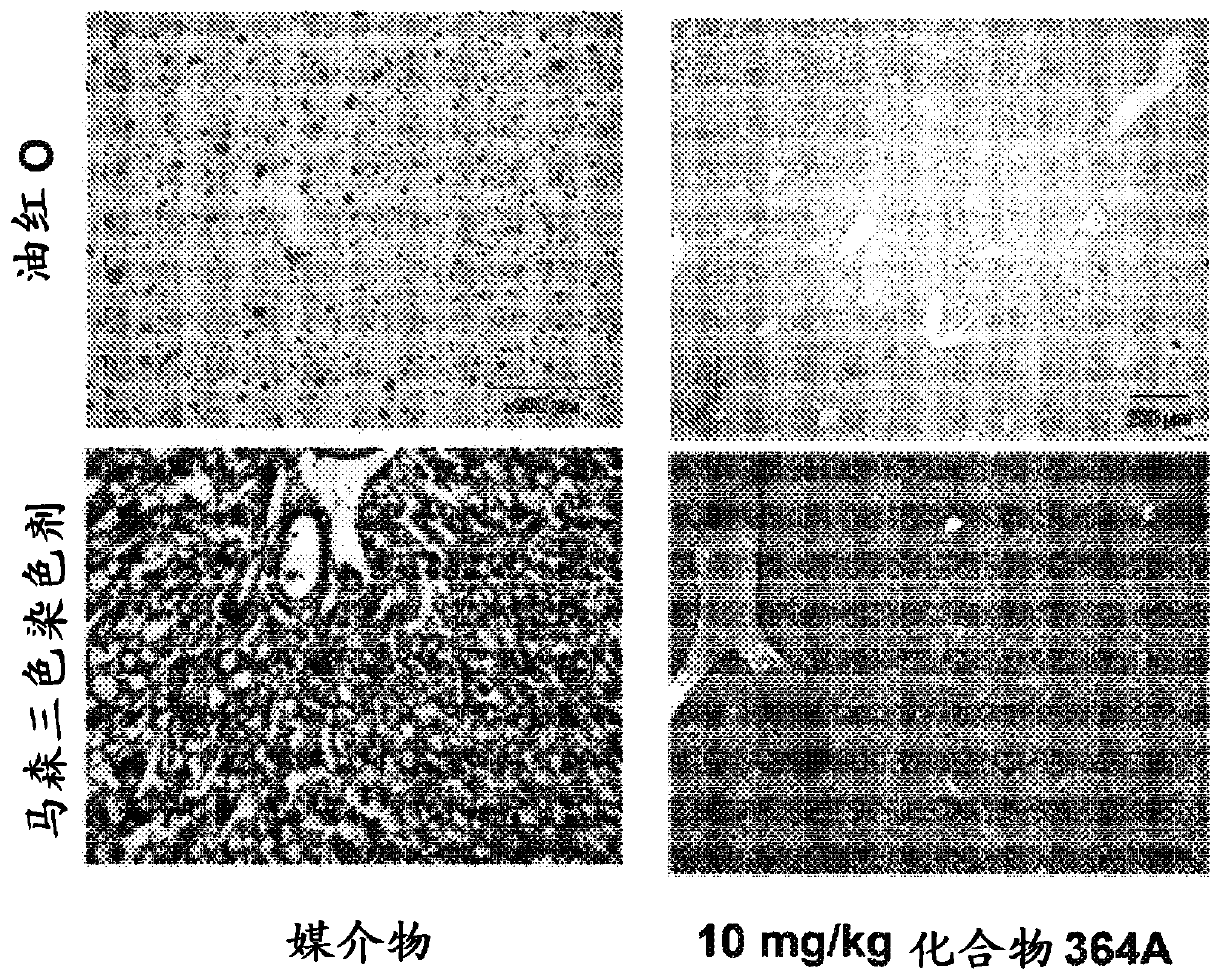
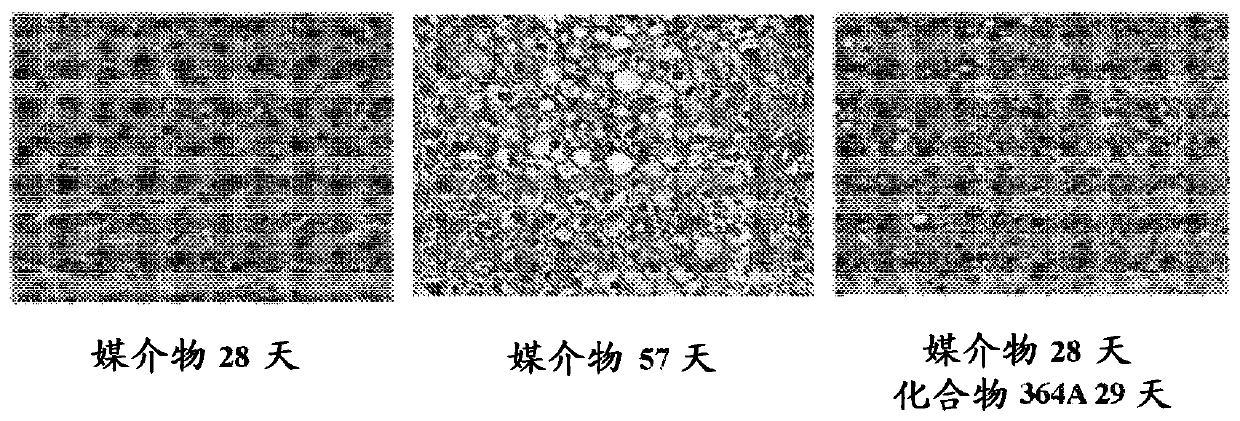
Examples
Embodiment 1
[5669] Example 1 - FASN Inhibition by Compounds of the Disclosure
[5670] Determination of FASN biochemical activity: FASN enzyme was isolated from SKBr3 cells. SKBr3 is a human breast cancer cell line with high expression levels of FASN. FASN is expected to comprise about 25% of the cytosolic protein in this cell line. SKBr3 cells were homogenized in a dounce homogenizer, followed by centrifugation at 4°C for 15 minutes to remove particulate matter. Supernatants were then analyzed for protein content, diluted to appropriate concentrations and used to measure FASN activity. The presence of FASN was confirmed by immunoblot analysis. A similar method for isolation of FASN from SKBr3 cells is described in Teresa, P. et al., (Clin. Cancer Res. 2009; 15(24), 7608-7615).
[5671] FASN activity of SKBr3 cell extracts was determined by measuring NADPH oxidation or the amount of thiol-containing coenzyme A (CoA) released during the fatty acid synthase reaction. The dye CPM (7-d...
Embodiment 2
[5672] Example 2 - Antiviral Activity
[5673] The antiviral activity of constructs (I-Z) was assessed using the HCV 1b replicon system:
[5674]
[5675] The replicon was constructed using the ET (luc-ubi-neo / ET) cell line, which is an HCV replicon with a stable luciferase (Luc) reporter and Three cell culture-adaptive mutant Huh7 human hepatoma cell lines (Pietschmann et al. (2002) J. Virol. 76:4008-4021). The HCV Replicon Antiviral Evaluation Assay investigated the effect of compounds at six half-log concentrations. Human interferon alpha-2b was included in each run as a positive control compound. Sub-confluent cultures of the ET line were plated out in 96-well plates dedicated to analysis of cell number (cytotoxicity) or antiviral activity and drugs were added to the appropriate wells the next day. Cells were processed 72 hours later when they were still near confluence. Determination of EC 50 (the replicon was inhibited by 50% and 90% respectively), IC 50 (conc...
Embodiment 3
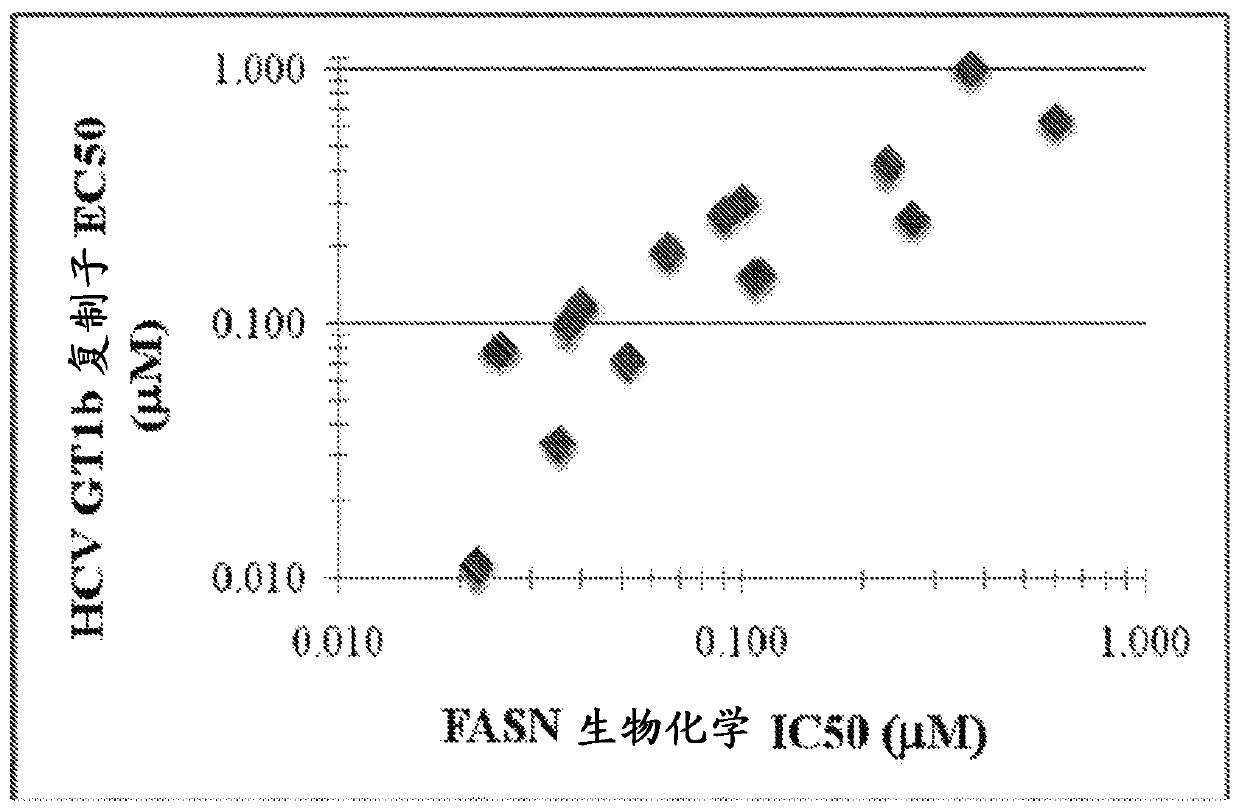
[5678] Example 3 - FASN Inhibition Correlates with HCV Inhibition
[5679] The antiviral activity of 15 compounds of the disclosure was measured using the HCV replicon system (numbers correlate to compounds in Table 1). According to published methods (Lohmann et al., (1999) Science 285(5424): 110-113; Lohmann et al., (2001) J. Virol 75(3): 1437-1449 and Qi et al., (2009) Antiviral Res .81(2):166-173), Huh7 was used to establish the replicon cell line 1b (HCV 1b / Luc-Neo replicon (1b Con1 integrated firefly gene)) by G418 selection. Replicon assembly using synthetic gene fragments. The GT1b line has PV-EKT and has 3 adaptive mutations E1202G (NS3), T1280I (NS3), K1846T (NS4B) and the backbone is Con1. The medium is:
[5680] DMEM supplemented with 10% FBS, G418 (250 μg / ml), streptomycin (100 μg / ml) / penicillin (100 U / ml), L-glutamine (100×), NEAA (100×)
[5681] Culture medium was prepared as follows:
[5682] 500 ml DMEM medium (Gibco, catalog number 11960-077)
[5683] ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com