A combination marker and its application in the preparation of gastric cancer risk prediction kit and its measurement system and method
A technology for detecting kits and markers, which is applied in biochemical equipment and methods, microbe determination/testing, biological testing, etc., and can solve problems such as the lack of combined markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051]The inventors used next-generation sequencing technology to obtain LIF, MIF, CCND2, BCL1L2, Expression of FABP1, ACSL5, ACADS, TBC1D1, CDX2 at gene level.
[0052] After analyzing the reference group of 38 cases of atrophic gastritis, the parameters of the formula are as follows:
[0053]
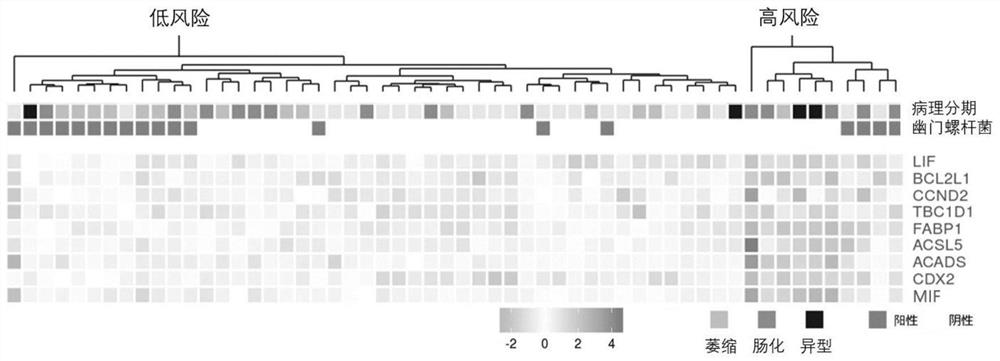
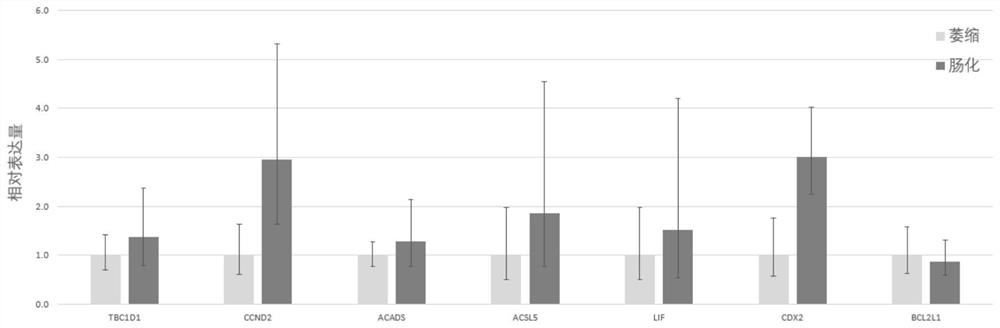
[0054] Based on the above parameters, the relative expression levels of each of the nine genes in 56 patients were calculated, and the risk grouping of the patients was obtained using the risk index calculation formula in the present invention, which was found to be significantly associated with the clinical stages of the patients (p=0.008457). And compared with the clinical stage, the expression levels of molecules such as LIF and FABP1 were more significantly correlated with the expression levels of CDX2 known to be related to intestinal metaplasia (such as figure 1 shown), suggesting that the combined markers in the present invention may be more closely related to the specific ...
Embodiment 2
[0057] Using the immunohistochemical detection kit according to the present invention to obtain seven molecular LIF, CCND2, BCL1L2, FABP1, ACSL5, The expression of ACADS and TBC1D1 at the protein level (MIF and CDX2 are not detected in this example, and can be directly replaced by zero value during calculation). The kit utilizes immunohistochemistry (IHC) to measure expression levels of the combined markers. 10% buffered formalin was used to fix paraffin-embedded surgical samples, and the tissue section was 4 μm / sheet.
[0058] The test kit in this embodiment includes the following components:
[0059] (1) Reagent A: blocking solution, which is 10% goat serum;
[0060] (2) Reagent B: diluted ready-to-use anti-LIF primary antibody;
[0061] (3) Reagent C: diluted ready-to-use anti-CCND2 primary antibody;
[0062] (4) Reagent D: diluted ready-to-use anti-BCL2L1 primary antibody;
[0063] (5) Reagent E: diluted ready-to-use anti-FABP1 primary antibody;
[0064] (6) Reagent ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com