Herbicidin biosynthetic gene cluster and application thereof
A gene and analog technology, applied in the field of Herbicidin biosynthesis gene cluster, can solve the problem that the biosynthesis gene cluster has not been reported.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
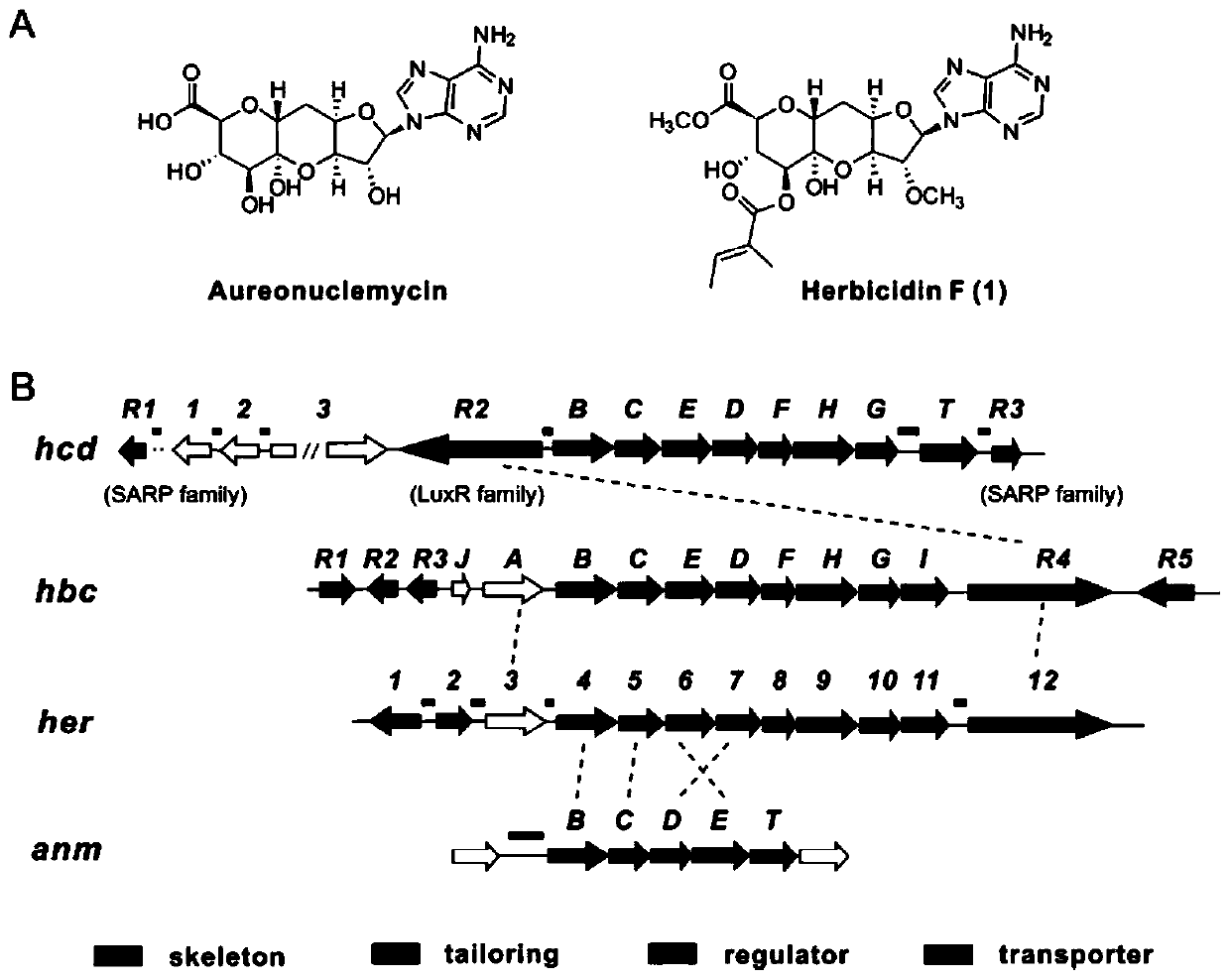
[0112] Example 1. Bioinformatics analysis and identification of herbicidin F biosynthetic gene cluster (hcd) in S. mobaraensis US-43
[0113] S. mobaraensis US-43 is a relatively mature strain (CPCC 204095) studied in our laboratory. Over the years, research on the metabolites of this strain has found that it can produce a large number of other types of compounds in addition to glycopeptide antibiotics. In order to systematically explore other types of metabolites, we sequenced the whole genome of S. mobaraensis US-43, and predicted that its genome contains at least 47 secondary metabolite biosynthesis gene clusters by using antiSMASH 4.0.0 software [20] . Through LC-MS analysis of the fermentation broth of the strain, compound 1 was detected as a trace component, and compound 1 was presumed to be herbicidin F by UV and MS / MS data analysis.
[0114] Although the structure of Herbicidin F has been determined, its biosynthetic pathway has not been reported yet. Compound aureon...
Embodiment 2
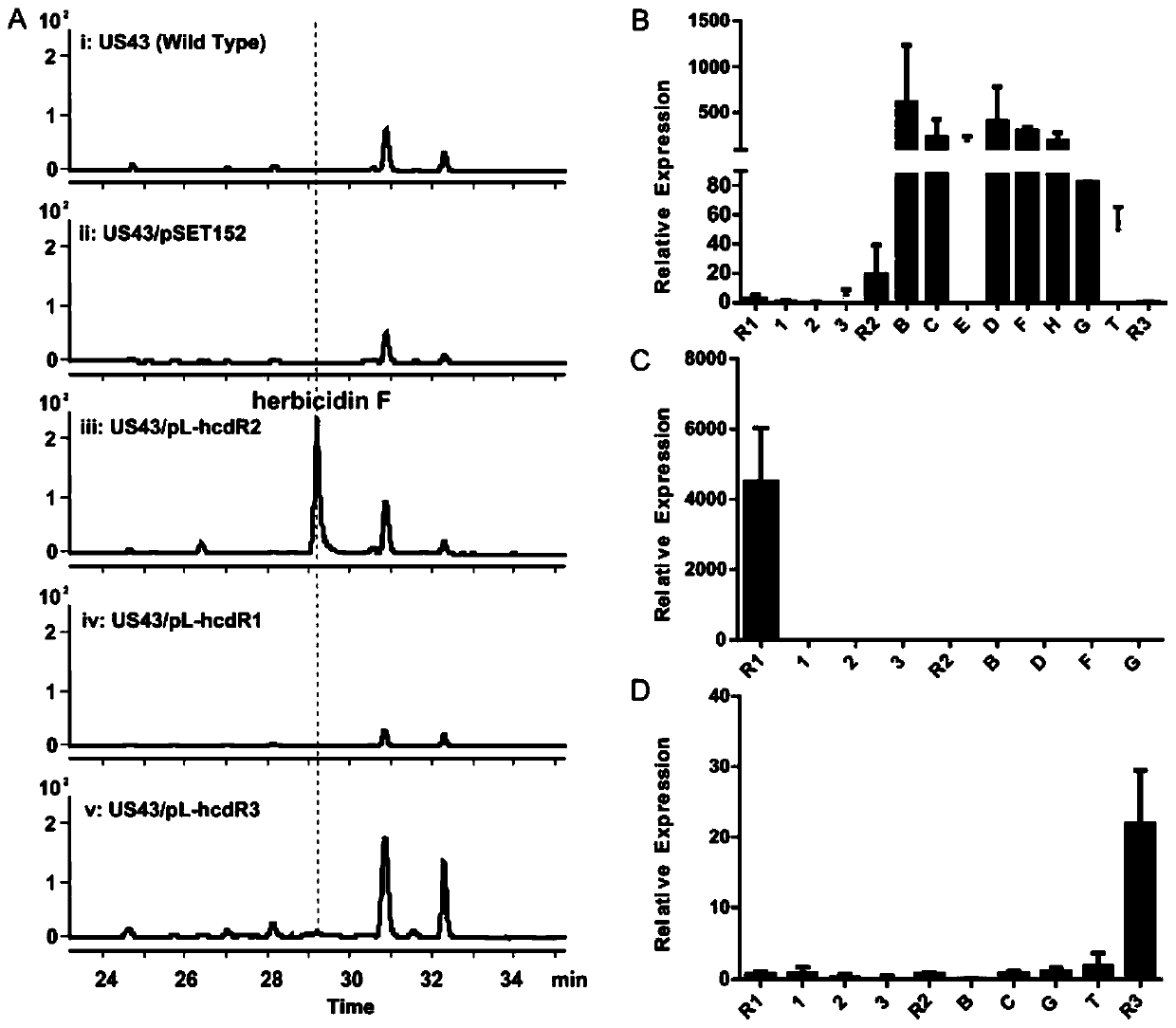
[0118] Example 2, Confirmation of pathway-specific regulatory proteins in hcd
[0119] The predicted hcd gene cluster contains three transcriptional regulatory genes hcdR1, hcdR2, and hcdR3. We constructed overexpressed strains of each transcriptional regulatory gene, and detected the changes in the yield of herbicidin F in the fermentation broth of the overexpressed strains by HPLC to judge the prediction Whether the gene cluster hcd is responsible for herbicidin F biosynthesis, and further determine whether hcdR1, hcdR2 and hcdR3 are pathway-specific regulatory genes in herbicidin F biosynthesis. We cloned each transcriptional regulator gene into pSET152 [21] The derived expression plasmid vector pL646 [22] Above, it contains the φC31attP site, which can be integrated into the attB site in the Streptomyces genome, and contains a strong promoter ermE*p and the SD sequence of the tuf1 gene upstream of the multiple cloning site. The constructed overexpression plasmids pL-hcdR...
Embodiment 3
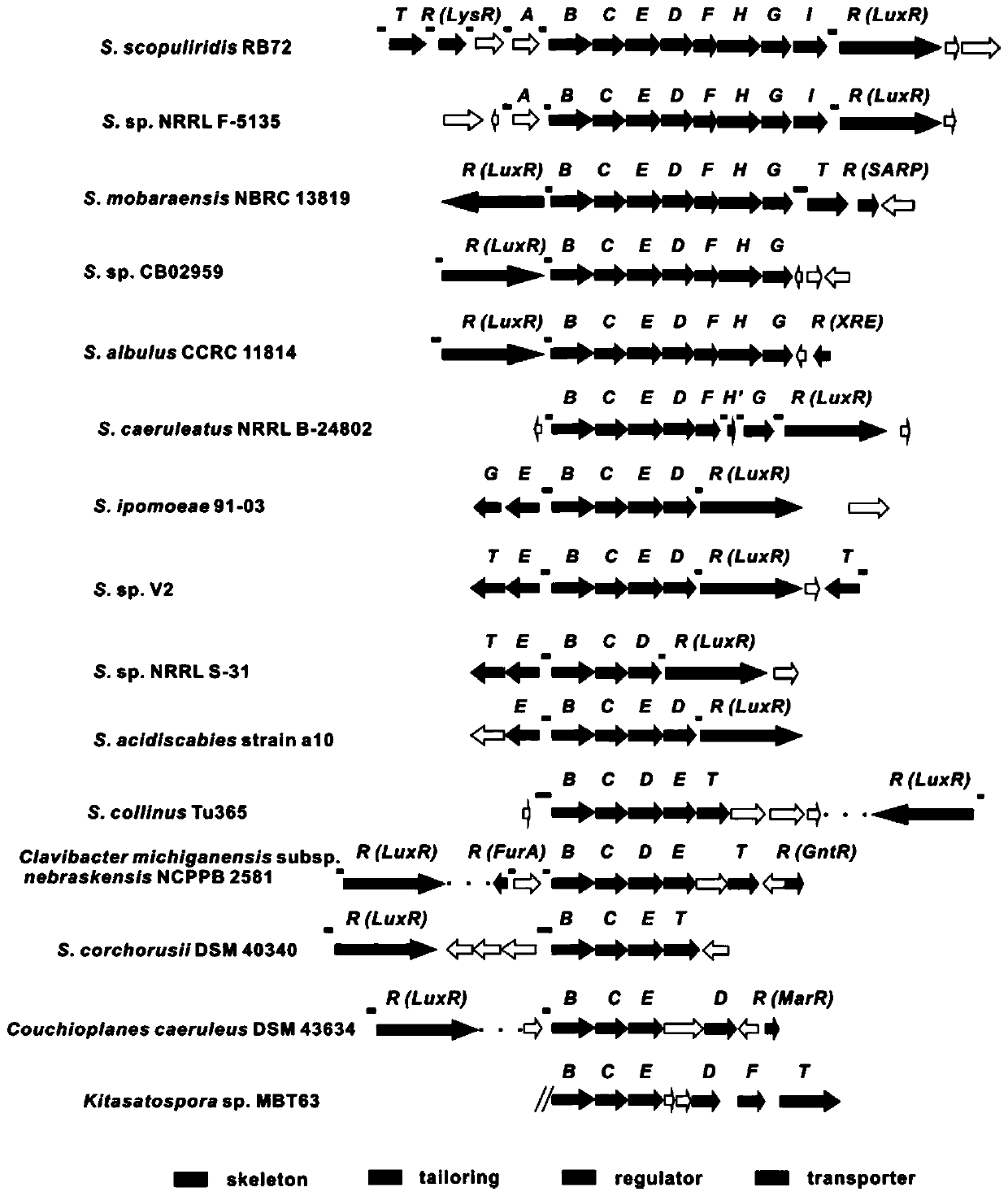
[0125] Example 3, HcdR2 is a relatively conserved pathway-specific regulatory protein in herbicidin biosynthesis
[0126] The pathway-specific regulatory protein HcdR2 belongs to the LuxR family. According to BLASTp analysis, there is a typical helix-turn-helix (HTH) DNA-binding domain (DBD) at the C-terminus of the regulatory protein. We searched for similar sequences in GenBank with hcdB / C / D / E as the target, and found that at least 15 actinomycetes contained hcd / hbc / her / anm similar gene clusters ( image 3 ), and 11 gene clusters contained transcriptional regulatory proteins belonging to the LuxR protein family. Among these LuxR family regulatory proteins, except for the LuxR protein in the gene cluster of strain S. mobaraensis NBRC 13819 (comparison analysis with HcdR2, 99.78% amino acid similarity), the rest of the LuxR regulatory proteins were compared with HcdR2, and the comparison results They were shown to have 30-43% amino acid similarity over the full length. HHpre...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap