Analog cancer genome sequencing data generating device
A genome sequencing and data generation technology, applied in sequence analysis, bioinformatics, informatics, etc., can solve problems that cannot be simulated, cannot be evaluated by software, and achieve performance evaluation and improvement effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
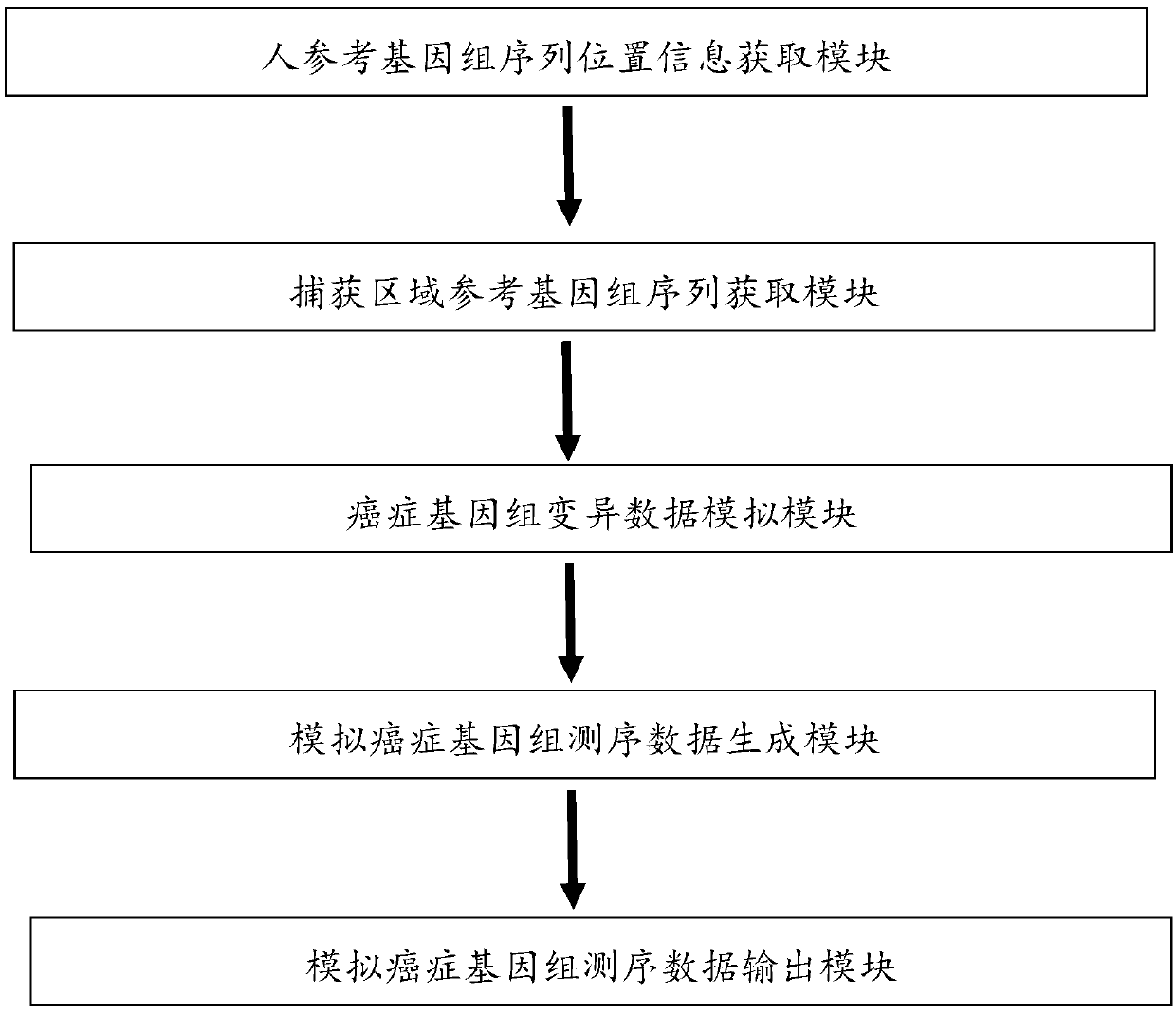
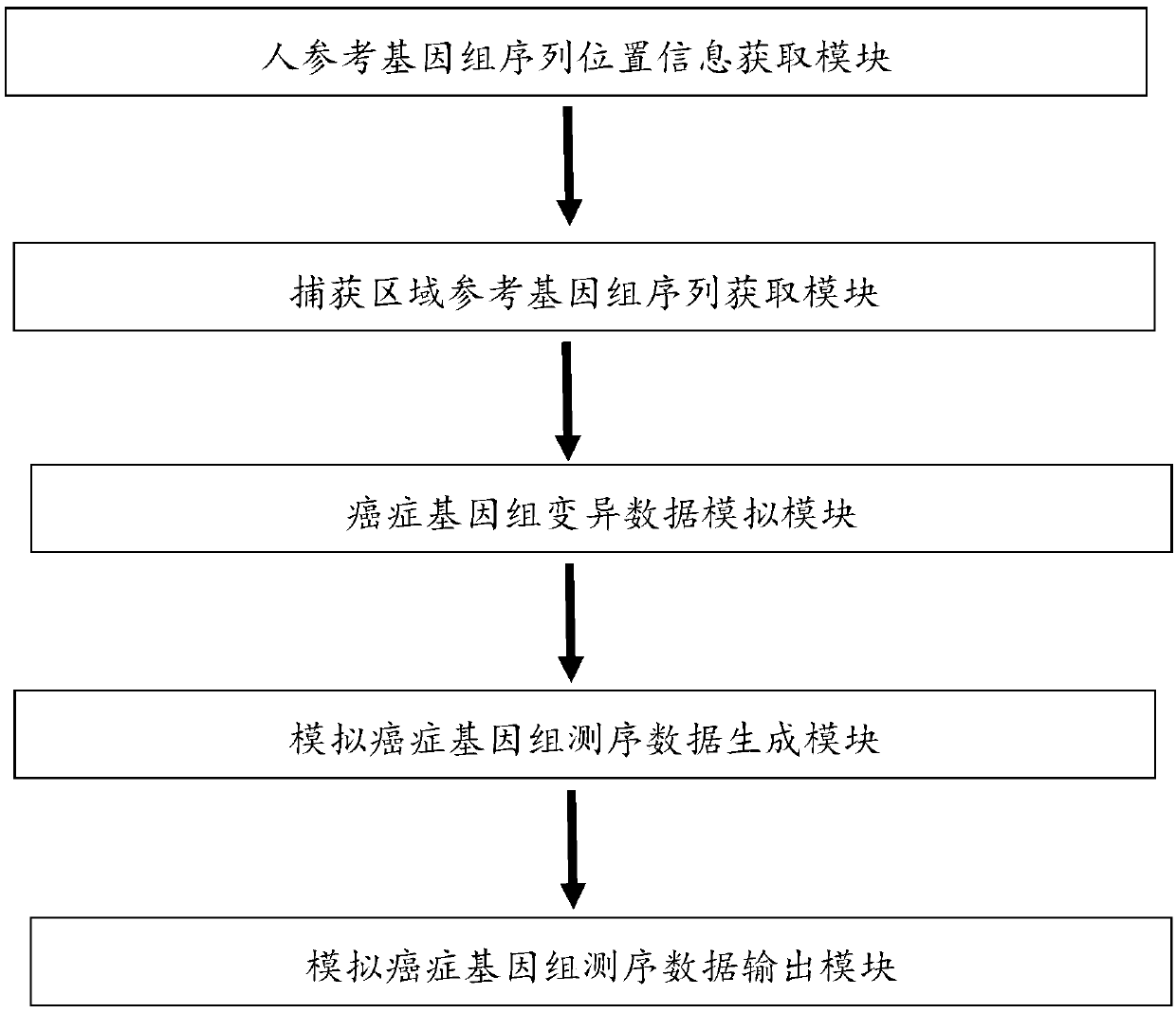
[0040] Target:
[0041] Generate simulated cancer genome sequencing data, simulate SNPs and INDELs with different frequency gradients, and use them to evaluate the performance of three variant detection software, GATK, mutect, and varscan.
[0042] step:
[0043] 1. The capture region location test.bed (254757bp) is randomly divided into 7 parts: respectively used to generate fastq files with mutation frequencies of 0.005, 0.01, 0.05, 0.1, 0.5, 0.9, and 1.
[0044] 2. Obtain the reference sequence fasta file corresponding to each location area file.
[0045] 3. Generate a variant fasta file (including 227 SNPs and 23 INDELs) on the reference sequence with snpfreq=0.001 and indfreq=0.001 / 10;
[0046] 4. With a depth of 1000 and a PE read length of 75, simulate and generate fastq files at each mutation frequency; merge the fastq files of each frequency.
[0047] 5. Get the bam file after comparing the fastq file, and then extract the bam file in the test.bed area
[0048] 6....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com