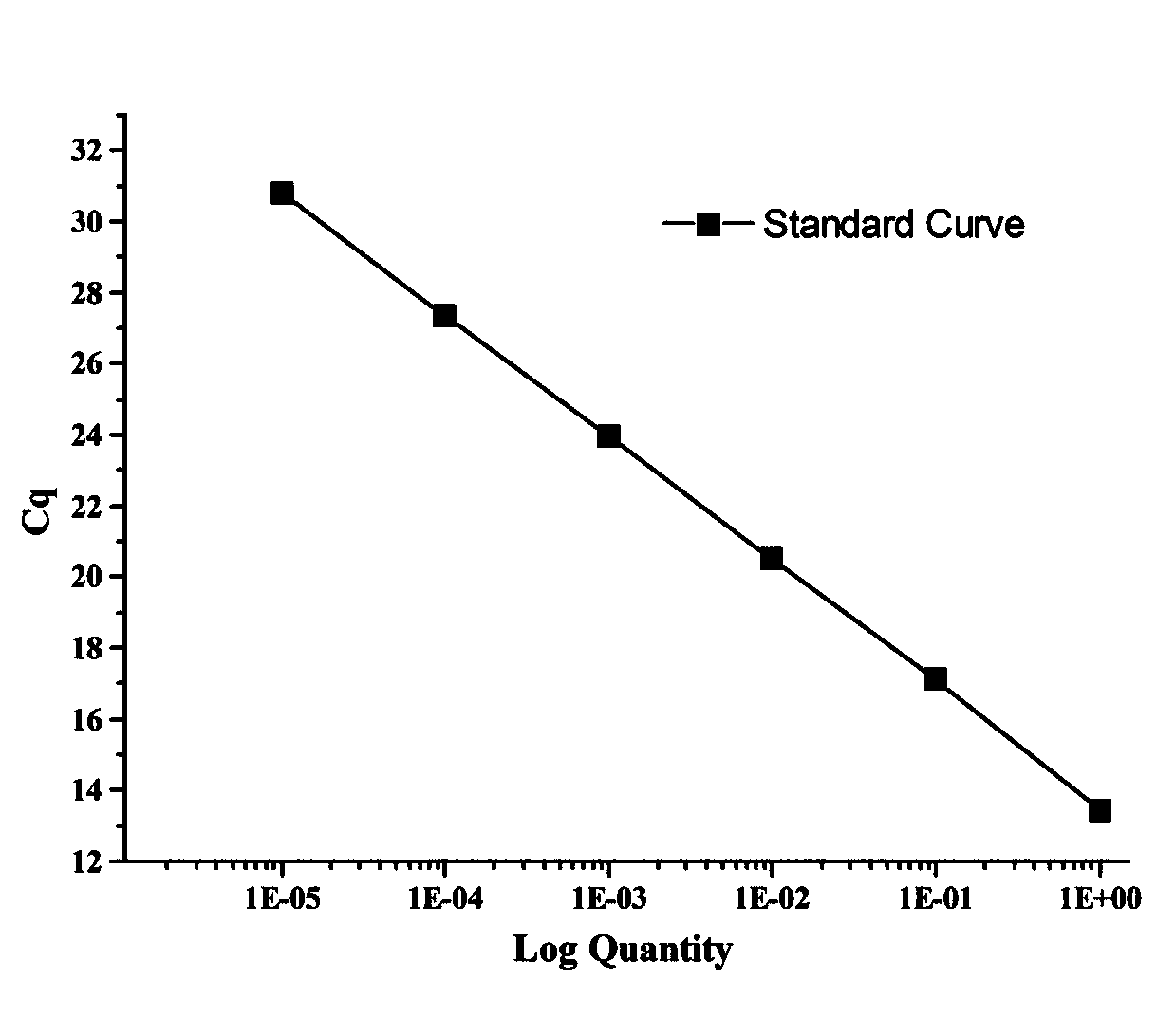
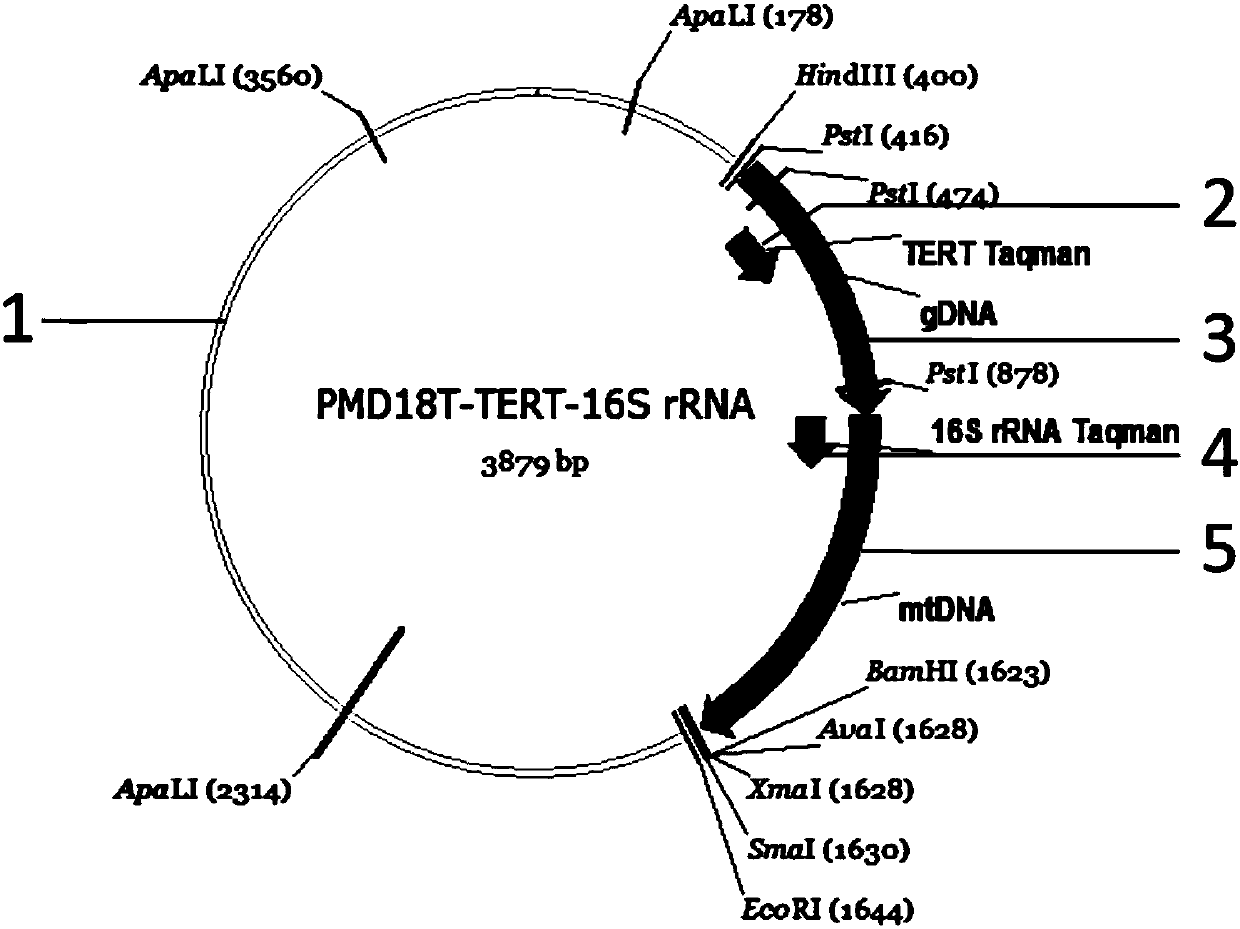
Method for quantitatively detecting nuclear genome copy number and human mitochondrial genome copy number by fluorescence
A mitochondrial genome and fluorescent quantitative detection technology, applied in the field of molecular biology, can solve the problems of difficult quantitative counting and inability to accurately reflect the copy number of mtDNA, and achieve the effects of reducing system errors, high specificity, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0019] This embodiment specifically includes the following steps:
[0020] Step 1, for the selected TERT gene and 16s_rRNA gene, design corresponding amplification primers, TERT gene clone amplification upstream primer (as shown in Seq ID No.2), specifically: 5'-CTCATTCCCACCCTTGAAATTGC-3', Reverse cloning amplification primer (as shown in Seq ID No.3), specifically: 5'-agcgGCTTCCTCAGGAACACCAAGAAG-3'; 16s_rRNA upstream cloning amplification primer sequence (as shown in Seq ID No.4), specifically: 5 '-aagcCGCTTTGACTGGTGAAGTCTTAGC-3', reverse cloning amplification primer (as shown in Seq ID No.5), specifically: 5'-CGTTCAAGCTCAACACCCACTAC-3', wherein the sequence of lowercase letters is a cohesive end, and these two PCR products can be Connected by secondary PCR, wherein the PCR reaction system for the first amplification is 30 μL, including 15 μL of 2×PCR reaction buffer (TOYOBO company), 1 μL of forward and reverse amplification primers with a concentration of 10 μmol / μL, 0.4 μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com