Epothilone biosynthetic gene transcriptional regulation protein and preparation method thereof
A technology of epothilone and gene transcription, which is applied in the field of genetic engineering and can solve the problems of high price and low yield of epothilone
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Embodiment 1: Cloning and analysis of promoter P3:
[0020] 1. Cloning of the promoter sequence
[0021] two,
[0022] 1. Design primers:
[0023] P3 upstream primer: 5'-TGGCGTCGGGCGCGGGG-3'
[0024] P3 downstream primer: 5'-TTGGGGATTGGAGAC-3'
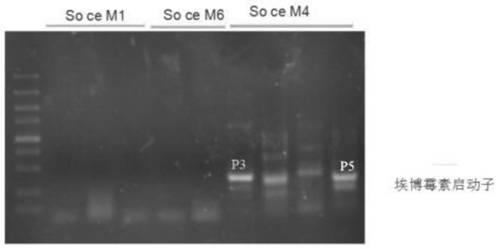
[0025] 2. Extract the genomes of three bacterial strains of S. cellulosus So ce M1, So ce M4 and So ce M6 with the bacterial genome extraction kit, according to the above p3 primer (comprising the P3 upstream primer and the P3 downstream primer) PCR, with Epothilium The P3 and P5 fragments ( figure 1 ), but the non-epothilone-producing S. cellulosus So ce M1 and So ceM6 could not amplify any fragments, indicating that the P3 and P5 fragments are specific fragments of epothilones producing bacteria.
[0026] The PCR reaction system is as follows:
[0027]
[0028] The PCR amplification procedure is as follows:
[0029]
[0030]
[0031] After recovery, the obtained PCR products were connected to pMD18-T vector, co...
Embodiment 2
[0032] Example 2: P3 promoter functional verification
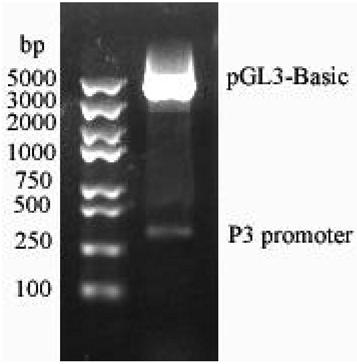
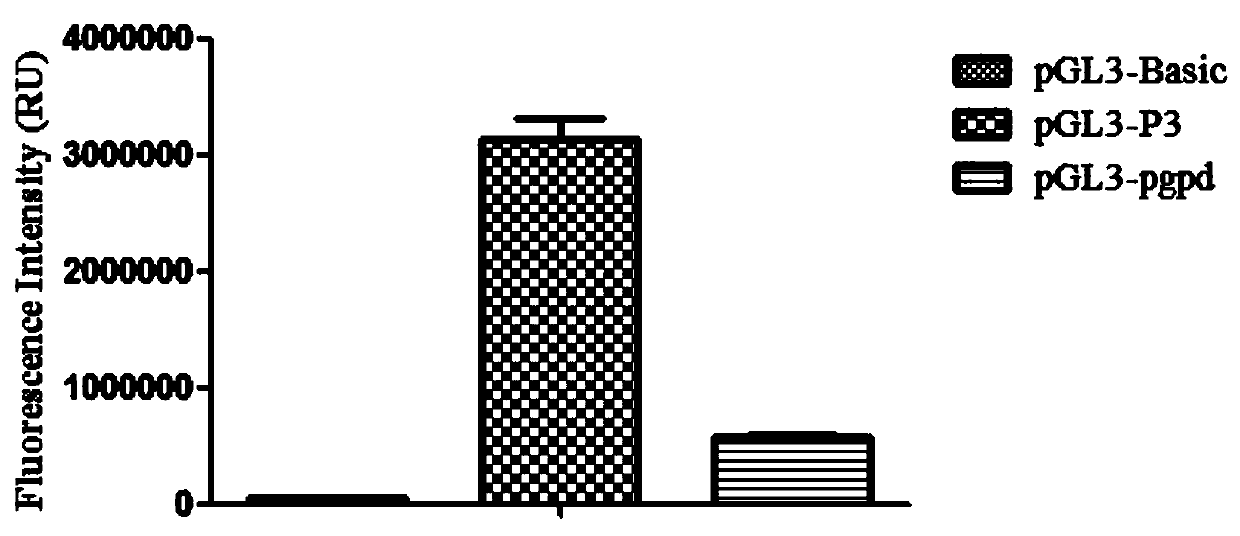
[0033] The P3 promoter was amplified by PCR, and XhoI and HindIII restriction sites were introduced at the 5' end and 3' end, respectively, recovered by cutting the gel, digested with XhoI and HindIII at 37°C for 4 hours, and recovered the product, and simultaneously used XhoI and HindIII37 Digest pGL3-Basic at ℃ for 2 hours, and recover the large fragment. Ligate the digested P3 promoter sequence and pGL3-Basic vector at 22°C for 3.0 h, transform into Escherichia coli DH5α, pick clones for expansion and culture, use P3 upstream primers and P3 downstream primers to carry out bacterial liquid PCR amplification respectively, and the obtained Positive clones were sequenced and verified, thus obtaining the PGL3-P3 recombinant vector (double enzyme digestion verification such as figure 2 shown), which is to insert the P3 promoter into the pGL3-Basic vector. The promoter pgpd was also inserted into the pGL3-Basic vector as d...
Embodiment 3
[0035]Embodiment 3: the acquisition of Sorangium cellulosum (Sorangium cellulosum) So ce M4 epothilone biosynthesis gene transcriptional regulatory protein, comprises the steps:
[0036] The 50 bp core sequence (TGCGATCTTGTGATTCCCCTTCTGATCTTTAAAATTTCCCGATCCCCCAT) of the promoter P3 of the epothilone biosynthesis gene cluster was labeled with the EMSA probe biotin labeling kit, and the specific steps were as follows:
[0037] Synthesize the P3 promoter core sequence and its complementary sequence, dilute to 1 μM with water, and configure the following system:
[0038]
[0039]
[0040] After mixing, incubate at 37°C for 30min, then mix equal volumes of sense strand and antisense strand, add annealing buffer (10×), mix well, and then perform annealing reaction under the following conditions: 95°C for 2 minutes, then 90 minutes Decrease 0.1°C every 8 seconds, and the resulting probe is used for EMSA detection.
[0041] The biotin-labeled P3 promoter was added to the strept...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com