Application of novel ScCas12a protein in nucleic acid detection
A nucleic acid technology, applied in the application field of new ScCas12a in nucleic acid detection, can solve the problem of inactivity of homologous proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1 Discovery of Cpf1 / ScCas12a gene
[0065] We found a Cas12a protein with DNA cutting activity from Smithella sp.SC_K08D17, denoted as ScCas12a (sequence shown in SEQ ID NO.1). The double-stranded DNA substrate, ScCas12a protein and gRNA were added to the reaction system in vitro, and it was found that the double-stranded DNA substrate could be specifically cleaved by the ScCas12a protein mediated by gRNA.
[0066] At the same time, the inventor's team found that when selecting the gRNA targeting sequence, the 5' end of the targeting sequence should have a 5'-TTTN-3' sequence, and the targeting sequence itself does not form a stable secondary structure, targeting sequence and The rest of the sequences do not form a stable secondary structure. Under this sgRNA design principle, ScCas12a has the activity of targeting DNA sequences in vitro or genome sequences in vivo for specific cleavage.
[0067] The following example gives the preparation of ScCas12a protein a...
Embodiment 2
[0068] Cloning and protein expression of embodiment 2 Cpf1 / ScCas12a gene
[0069] 1. PCR amplification of Cas12a sequence
[0070] (1) Design primers
[0071] The upstream and downstream primers were designed according to the ScCas12a sequence, and the sequence is as follows:
[0072] Upstream primer (shown in SEQ ID NO.4):
[0073] GTGCGGCCGCCATGCAGACCCTGTTTGAGAACTTCACA;
[0074] Downstream primer (shown in SEQ ID NO.5):
[0075] GTGGCGCGCCTGGCATAGTCGGGGACATCATATG.
[0076] (2) PCR amplification
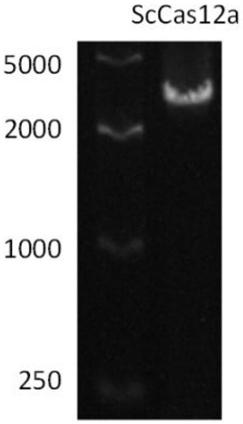
[0077] Utilize the above-mentioned upstream and downstream primers, and use high-fidelity DNA polymerase (phusion DNA polymerase) to perform PCR amplification on the target fragment at different annealing temperatures. The results are attached figure 1 Shown, PCR target band (about 4000bp).
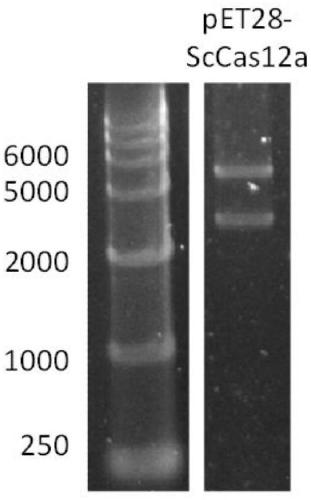
[0078] 2. Construction of recombinant plasmid pET-28a-ScCas12a
[0079] (1) PCR amplification product purification: the product after PCR amplification is purified by Qiagen's purifica...
Embodiment 3
[0093] Example 3 Purification of ScCas12a protein
[0094] 1. Purification method of ScCas12a protein
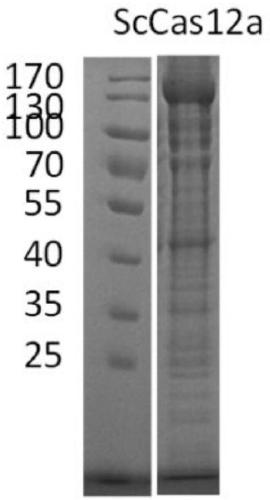
[0095] The bacteria solution after induction of expression was centrifuged, the bacteria were resuspended in the lysis buffer, ultrasonicated (70% amplitude, 2s On / 4s Off, 3 minutes, Sonics 750w ultrasonic instrument), and the supernatant was separated by centrifugation. Load protein lysate supernatant to equilibrated Ni Sepharose FF, wash away impurity proteins with lysis buffer greater than 30 times column bed volume, and elute with elution buffer, use Superdex 200, Tricorn 10 / 300 gel column for purification. After elution, SDS-PAGE analysis and observation results and gel column purification were carried out to obtain the purified Cas12a protein. Wherein, the lysis buffer contains 50 mM Tris-HCl, pH 8.0 300 mM NaCl, 5% glycerol, and 20 mM imidazole. Elution buffer contained 50 mM Tris-HCl, pH 8.0 300 mM NaCl, 5% glycerol, 250 mM imidazole.
[0096] The resulting prote...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com