Method for analyzing impurities of oligonucleotide sequence based on high-throughput sequencing and application
An oligonucleotide, high-throughput technology, applied in biochemical equipment and methods, microbial measurement/testing, library creation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
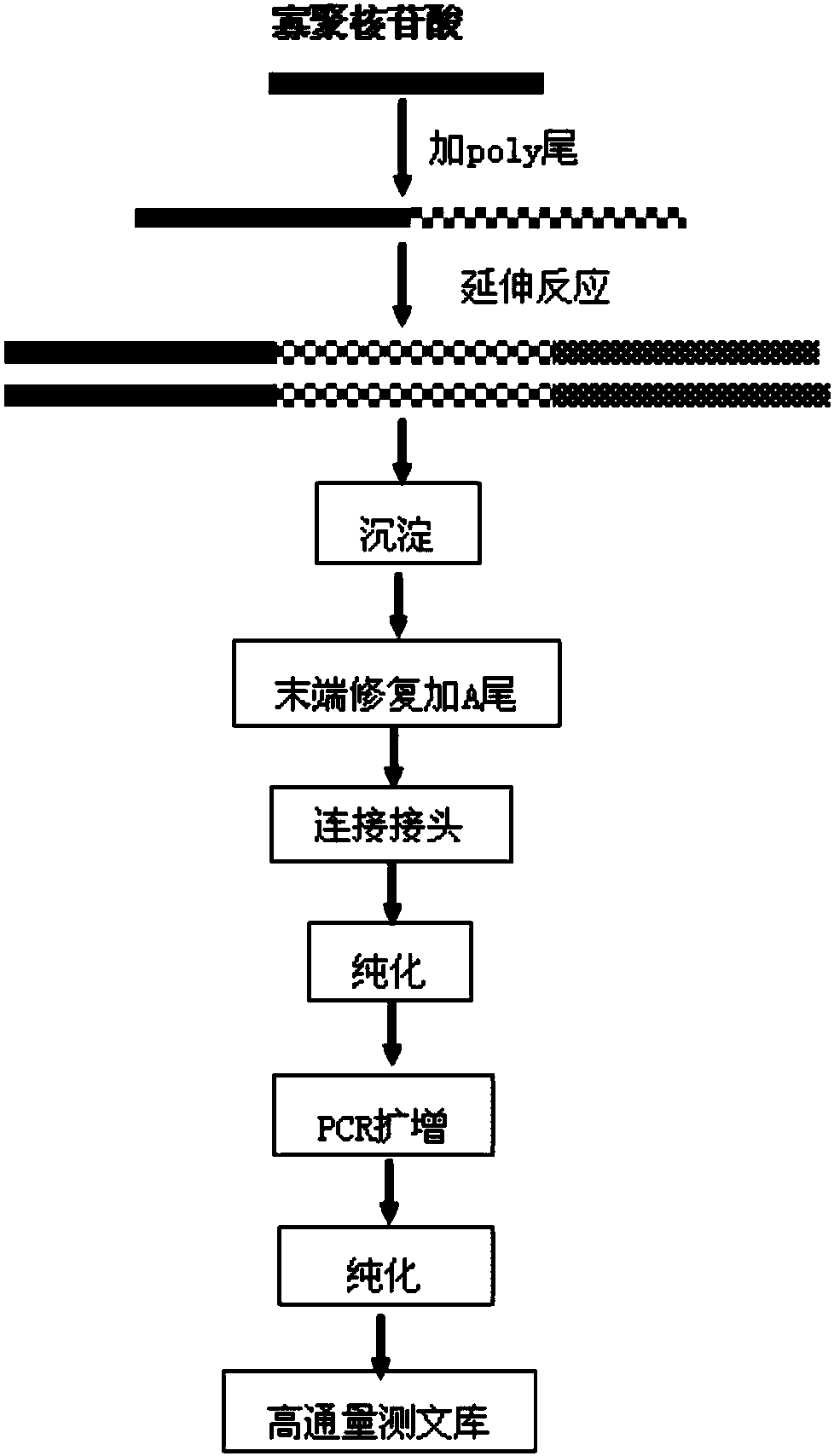
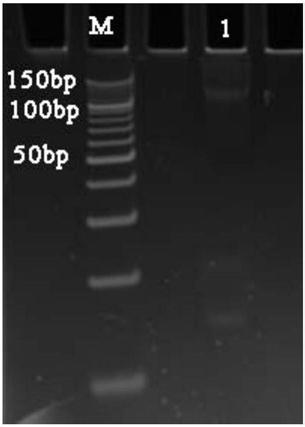
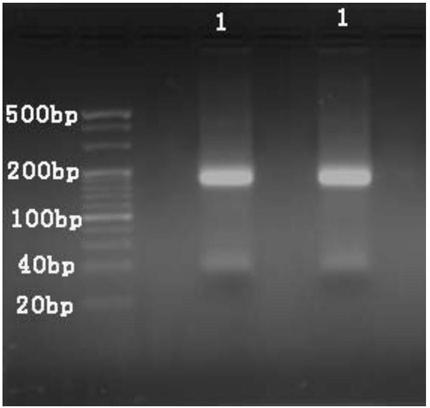
[0063] Example 1. A method for analyzing oligonucleotide sequence impurities based on high-throughput sequencing
[0064] 1. Oligonucleotide plus poly tail
[0065] 1. Add a poly A tail to the 3' end of the oligonucleotide sequence, and prepare a tailing reaction system according to the reagents and dosage in Table 1. The oligonucleotide sequence was as follows: 5'-CAGAGCAGCTTGTCTTTTCTTC-3' (SEQ ID NO: 5). Oligonucleotide sequences were synthesized by Shanghai Jierui Bioengineering Co., Ltd.
[0066] Table 1 is the tailing reaction system
[0067] Reagent
Amount added (μL)
13
Oligonucleotides (100 μM)
1
dATP (25μM)
0.5
5×TdT Buffer
4
terminal transferase
1.5
total capacity
20 μL
[0068] 2. React at 37°C for 25 minutes.
[0069] 3. React at 70°C for 10 minutes to inactivate terminal transferase.
[0070] 2. Reverse extension and amplification
[0071] 1. Add the reagents sh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com