Codominant Molecular Marker and Application of Rice Sub1 Sub1 Submergence Tolerance Gene
A molecular marker, co-dominant technology, applied in biochemical equipment and methods, DNA/RNA fragments, microbial determination/inspection, etc., can solve problems such as limiting the breeding efficiency of flood-tolerant rice varieties, and achieve convenient and fast detection procedures. , low cost, high detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Development of Sub1 Gene Co-dominant Molecular Marker
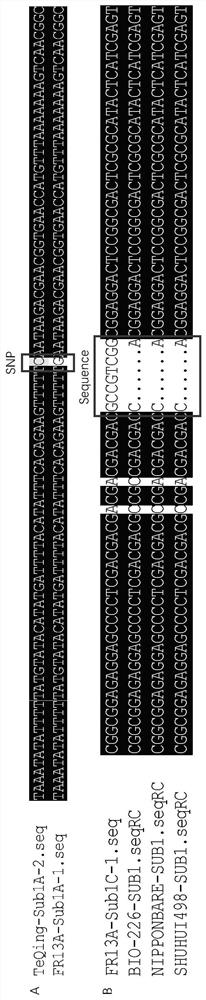
[0037] The rice Sub1 gene is located on chromosome 9. It is a gene cluster composed of Sub1A, Sub1B and Sub1C. Currently, it is known that Sub1A, Sub1B and Sub1C have 2 (or missing), 9 and 7 alleles respectively: SubA1 ~2, Sub1B1~9, Sub1C1~7, rice flood tolerance traits are mainly controlled by Sub1A-1, and Sub1A-1 only appears in the combination with Sub1C-1, therefore, by analyzing the Sub1A and Sub1C in different rice materials The sequence was amplified and sequenced, and DNAMAN and other software and NCBI database and other sequence analysis tools were used for local and online sequence comparison analysis, and a SNP site was obtained in the Sub1A gene interval, in which the flood tolerance allele Sub1A-1 was G, Allele Sub1A-2 of intolerance to flooding is C (eg figure 1 A); A specific nucleotide sequence was found in the Sub1C gene interval, wherein the Sub1C-1 allele was 5'-GCCGTCG-3', and the res...
Embodiment 2
[0038] The detection primer (ARMS-PCR primer) design of embodiment 2Sub1 gene co-dominant molecular marker
[0039] For the two Sub1 gene-specific molecular primers developed in Example 1, ARMS-PCR detection primers were designed, and the principles of primer design are as follows:
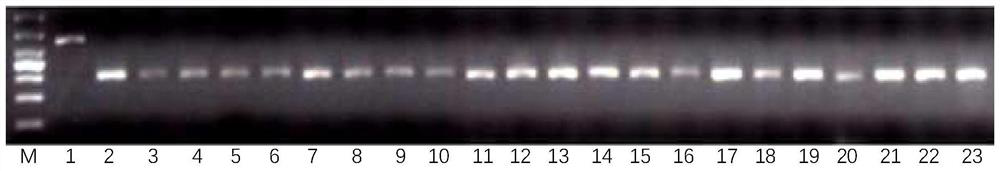
[0040] First, design primers for specific amplification of the Sub1A-1 allele for the specific SNP site in the Sub1A gene, use the C base complementary to the G base in the SNP as the 3' end of the primer to design the reverse primer Sub1A-R, and combine 3 The second base at the ' end is changed to G, and then the forward primer Sub1A-F paired with it is designed upstream, and the primers Sub1A-R and Sub1A-F (SEQ ID NO:2-3) can only be specifically amplified The Sub1A-1 allele type produced a 586bp band, while the Sub1A-2 allele type and the Sub1A gene deletion type amplified no band.
[0041] Secondly, design specific amplification intolerance allelic primers for the specific nucleotide sequence...
Embodiment 3
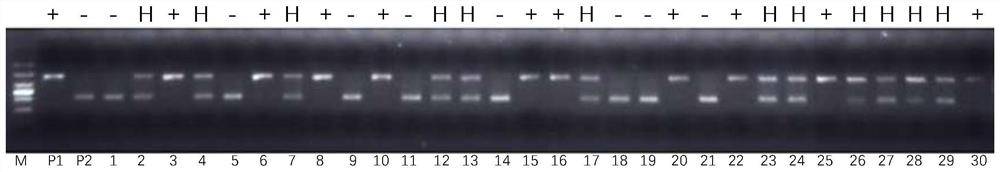
[0045] Example 3 Establishment of detection method for rice sub1 specific molecular marker for submergence tolerance gene
[0046] According to the detection primers of two molecular markers of Sub1 designed in Example 2, the reaction program and reaction system of PCR were designed, and through continuous optimization, the following reaction program and system were determined:
[0047] PCR reaction system (10 μL): 10×PCR reaction buffer 1 μL, 10 mM dNTP 0.8 μL, 4 primers (10 μM) 0.15 μL, 0.1 μL Taq DNA polymerase (2.5U / ul); 2 μL DNA template, double-distilled Water to make up the balance.
[0048] The PCR reaction conditions were: pre-denaturation at 95°C for 5 minutes; denaturation at 95°C for 30 seconds, annealing at 58°C for 30 seconds, extension at 72°C for 45 seconds, a total of 35 cycles; extension at 72°C for 8 minutes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com