Exosome loading ABCA1 mRNA and construction method and application thereof
A construction method and technology for exosomes, which are applied in the field of exosomes loaded with ABCA1 mRNA and their construction, can solve the problems of increasing the expression of ABCA1 protein in the liver, the inability to load ABCA1, and the large molecular weight, and achieve the effect of improving efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] (1) After the 3' end of the CDS sequence (nucleotide sequence shown in SEQ ID No.1) of ABCA1, a specific binding sequence AREs of the RNA binding protein HuR (shown in SEQ ID No.2) is fused Nucleotide sequence), namely ABCA1-AREs gene (nucleotide sequence shown in SEQ ID No.3), ABCA1-AREs gene was synthesized according to the nucleotide sequence shown in SEQ ID No.3.
[0085] (2) Using pcDNA3.1(-) as the backbone, cut the pcDNA3.1(-) vector with the restriction endonuclease EcoRV, mix and connect the linear vector obtained after digestion with the synthetic ABCA1-AREs gene, and screen , get pcDNA3.1-ABCA1-AREs expression vector.
[0086] (3) Trizol extracted the total RNA of mouse muscle cells; the total RNA was reverse-transcribed using Promega M-MLV reverse transcriptase to obtain mouse fibroblast cDNA:
[0087] The reverse transcription system is: 2 μg of RNA template, 4 μl of 5×RT buffer, 2 μl of dNTP mixture (10 mM), 2 μl of 0.1MDTT and 1 μl of M-MLV reverse trans...
Embodiment 2
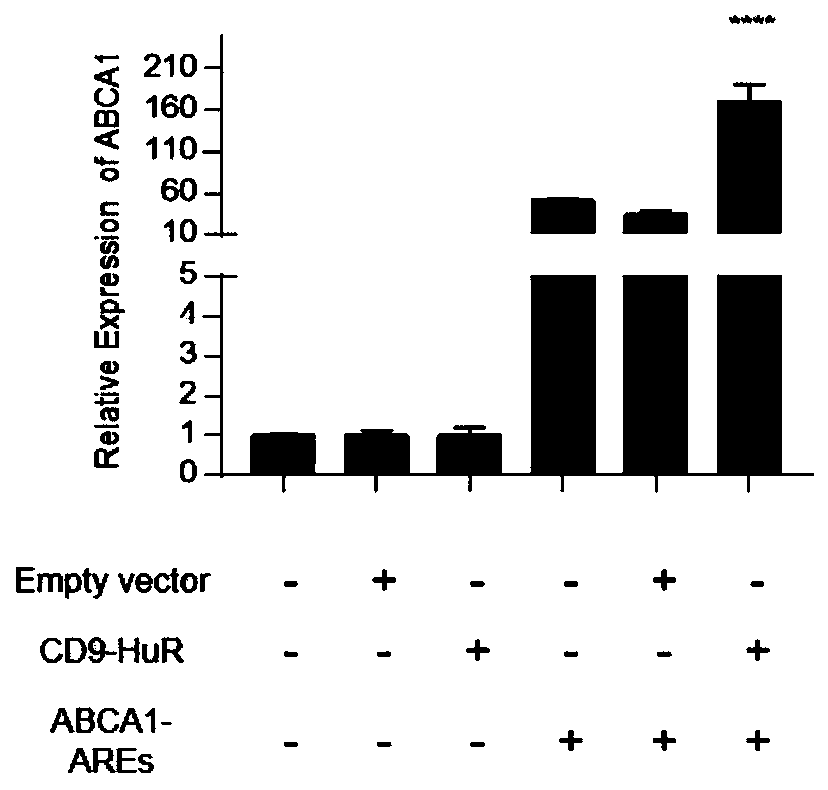
[0106] This experiment verified the efficiency of ABCA1 mRNA loading exosomes:
[0107] Detection method: Exosomes were lysed by Trizol, RNA was extracted according to the instructions of the RNA extraction kit, glycogen was added to precipitate RNA according to the instructions during the extraction process, followed by reverse transcription, and then qPCR was used to detect the loading efficiency of target RNA molecules.
[0108] The formula for calculating the relative expression of ABCA1 mRNA was as follows: β-actin was selected as an internal reference gene, and 2-ddCt was used to calculate the relative expression of ABCA1 mRNA in each exosome.
[0109] Sample: Exosomes expressed by hepatocytes that were not transfected with any vector were used as a control (Empty vector); the pcDNA3.1-CD9-HuR expression vector was prepared according to steps (3) to (6) described in Example 1; The pcDNA3.1-ABCA1-AREs expression vector was prepared by steps (1)-(2) described in Example 1....
Embodiment 3
[0113] 100 μL of exosomes prepared in Example 1 was injected into C57 mice through the tail vein, and after 48 hours, the mice were sacrificed to further collect liver tissue. At the same time, PBS was used as the control group, which was the same as the above operation.
[0114] Using WB to detect the expression of ABCA1 and the control group in the liver, the primers used are shown in Table 1, and the results are as follows figure 2 shown.
[0115] Table 1 Reaction primers used in WB detection
[0116]
[0117]
[0118] Wherein β-actin is an internal reference, PBS represents the expression level of ABCA1 in the liver of mice treated in the control group, and Exo-ABCA1-AREs represents the expression level of ABCA1 in the liver of mice injected with exosomes prepared in Example 1. Depend on figure 2 It can be seen that the exosomes loaded with ABCA1 mRNA constructed in the present invention significantly increased the expression of ABCA1 in mouse liver cells.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com