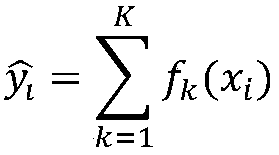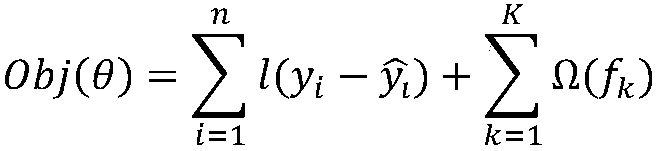Xgboost-based whole-genome RNA secondary structure prediction method
A secondary structure, whole genome technology, applied in the field of bioinformatics research, can solve the problems of many output targets and affecting the accuracy rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0022] The present invention will be further described in conjunction with the accompanying drawings.
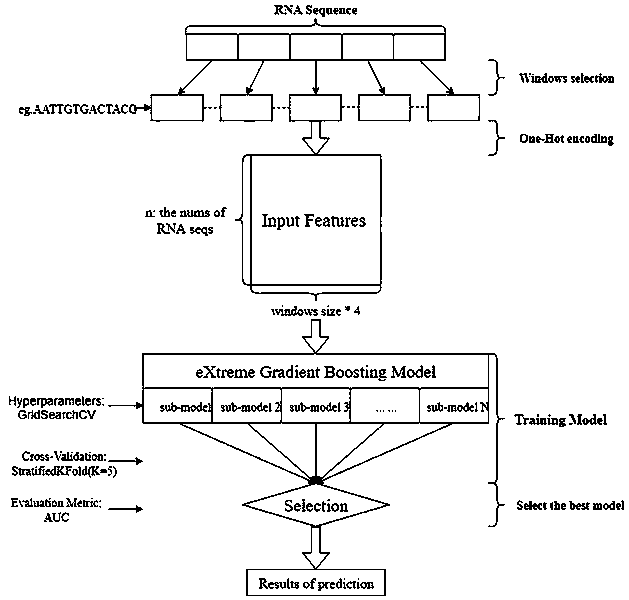
[0023] This method hopes to obtain the key features of the secondary structure formed by the primary sequence through supervised learning of the most basic sequence structure, and assist in judging how the secondary structure is formed from the primary sequence structure.
[0024] Such as figure 1 As shown, first obtain the dataset. The data set is derived from the results of biological experiments, and the possibility value of pairing between the RNA sequence and the base site in the RNA sequence is obtained from the results of the biological experiment. The datasets used include three, labeled as PARS-human, PARS-yeast, and PDB-Xray datasets. Specifically, the full name of PARS is "Parallel Analysis of RNA Structure (PARS)", which is an experimental method for determining the secondary structure of RNA through biological experiments proposed in 2010. PARS-Human refers to a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com