A multi-factor co-evolutionary method to increase the production of Cryptidinosa dha
A co-evolutionary, Cryptodinomorphic technology, applied in the directions of microorganism-based methods, methods using microorganisms, biochemical equipment and methods, etc., can solve problems such as inhibiting the growth of Cryptodininos cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
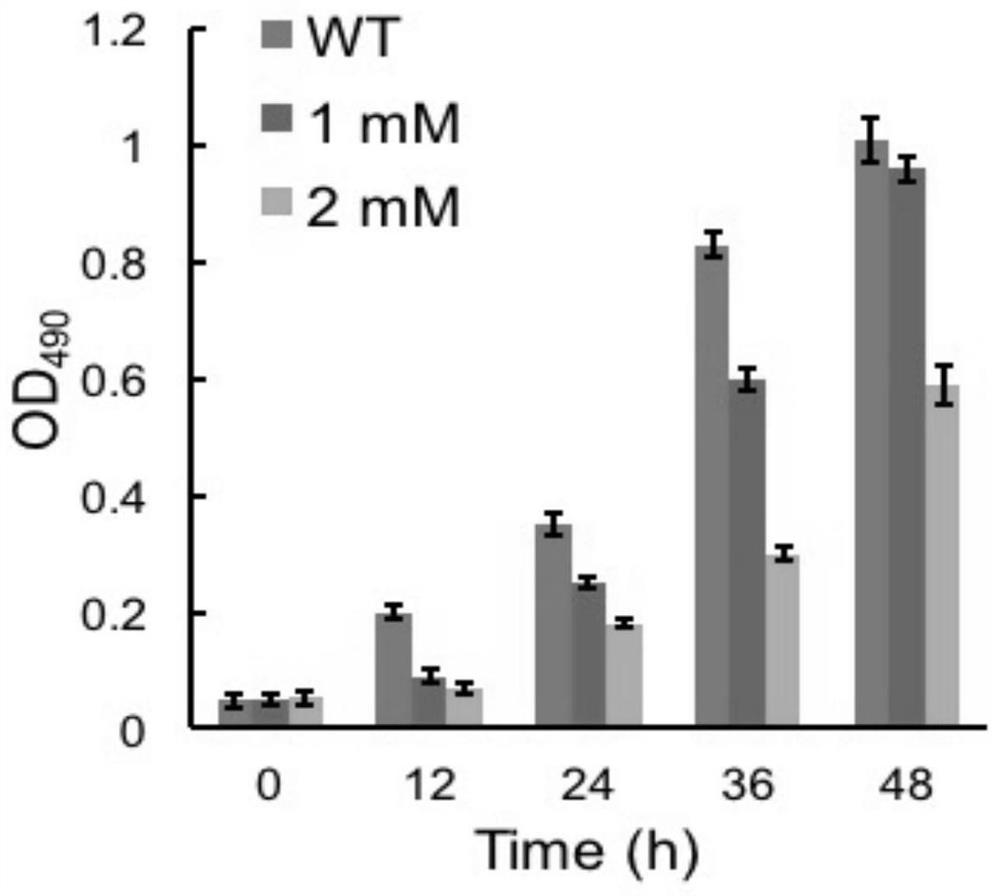
[0044] The first round of directional domestication using sethoxydim to provide selective pressure
[0045] 1. Prepare the C9N2 solid plate culture medium (5μM-20μM) with gradient concentration of sethenhexidine.
[0046] 2. Select 50 μL of fresh cells with an OD490nm of 0.8 and spread evenly on the resistant plate, and culture them upside down for about 2 weeks to observe the growth and determine the initial concentration for acclimation.
[0047] 3. Using C9N2 medium as the medium for continuous passage, wherein the initial concentration of sethenoxydim is 10 μM, insert 1 mL of fresh wild-type cells with an OD490nm of 0.8, and culture to the logarithmic phase.
[0048] 4. Transfer the bacterial strain obtained in the previous step into fresh C9N2 containing the same concentration of sethenoxydim, culture it to the mid-log phase and continue to subculture until the Cryptodinoflagellate cells are no longer sensitive to 10 μM sethenoxydim.
[0049] 5. The strains obtained in t...
Embodiment 2
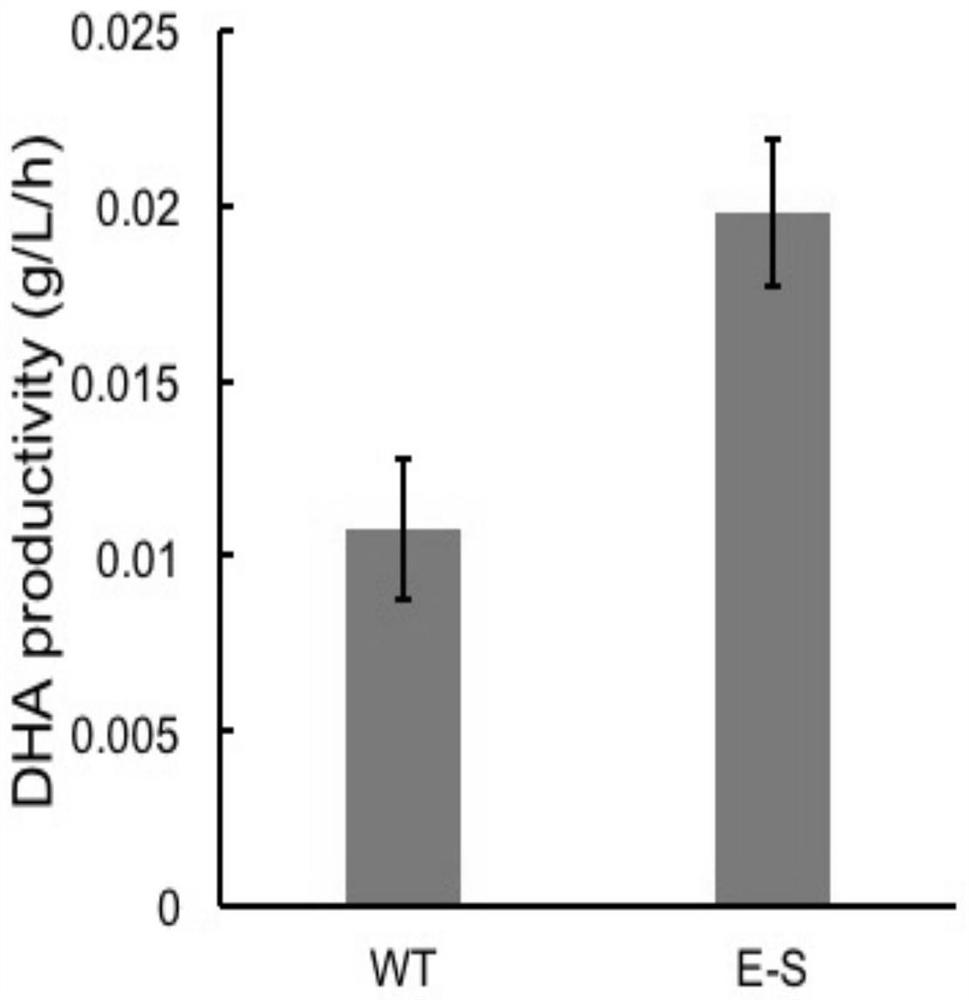
[0054] This example is basically the same as Example 1, except that the initial strain used is the mutant strain obtained in Example 1, and sesamol is used to provide selection pressure for the second round of directional domestication.
[0055] After the above two examples, the obtained dual-acclimated bacterial strain is desensitized to sesamol. Therefore, during the fermentation process of the double domesticated strain, by adding 1 mM sesamol, the yield of DHA increased by about 130% compared with the wild type.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com