A method for functional prediction of single nucleotide genomic variation in a non-coding region
A single nucleotide variation, single nucleotide technology, applied in the field of genes, to achieve the effect of low time and economic cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
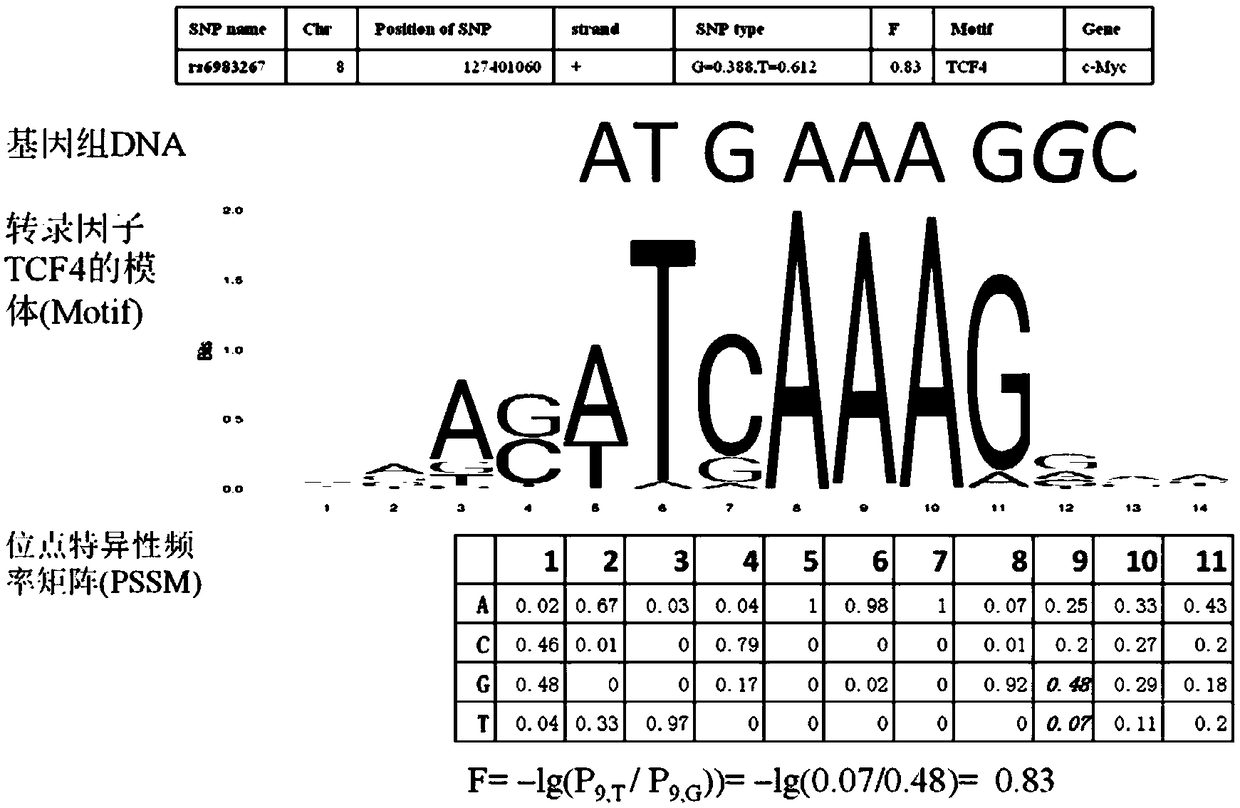
[0031] Embodiment 1: Accuracy experiment of the prediction method of the present invention: Calculating the influence of a single nucleotide polymorphism of DNA on a transcription factor binding site on transcription factor binding
[0032] The proto-oncogene c-MYC has a regulatory region (8q24) at a distance of 335kb. This region has a SNP (rs6983267) (genome assembly version GRCh37.p13). In the Thousand Genomes Project, 1008 Asian populations (Phase3_V1-EAS ), on the positive strand, the frequency of this site is guanine (G) is G=0.388, and the frequency of cytosine (C) is C=0.612. In the European population (thousand genomes, population size 1006), the frequency was G=0.499, T=0.501. The DNA sequence ("ATGAAAGGC") where the SNP is located is the binding site of the transcription factor TCF4, and the target gene regulated by it is c-MYC.
[0033]In the European population, genotype G has a slightly lower frequency (0.501) and genotype T has a slightly higher frequency (0.49...
Embodiment 2
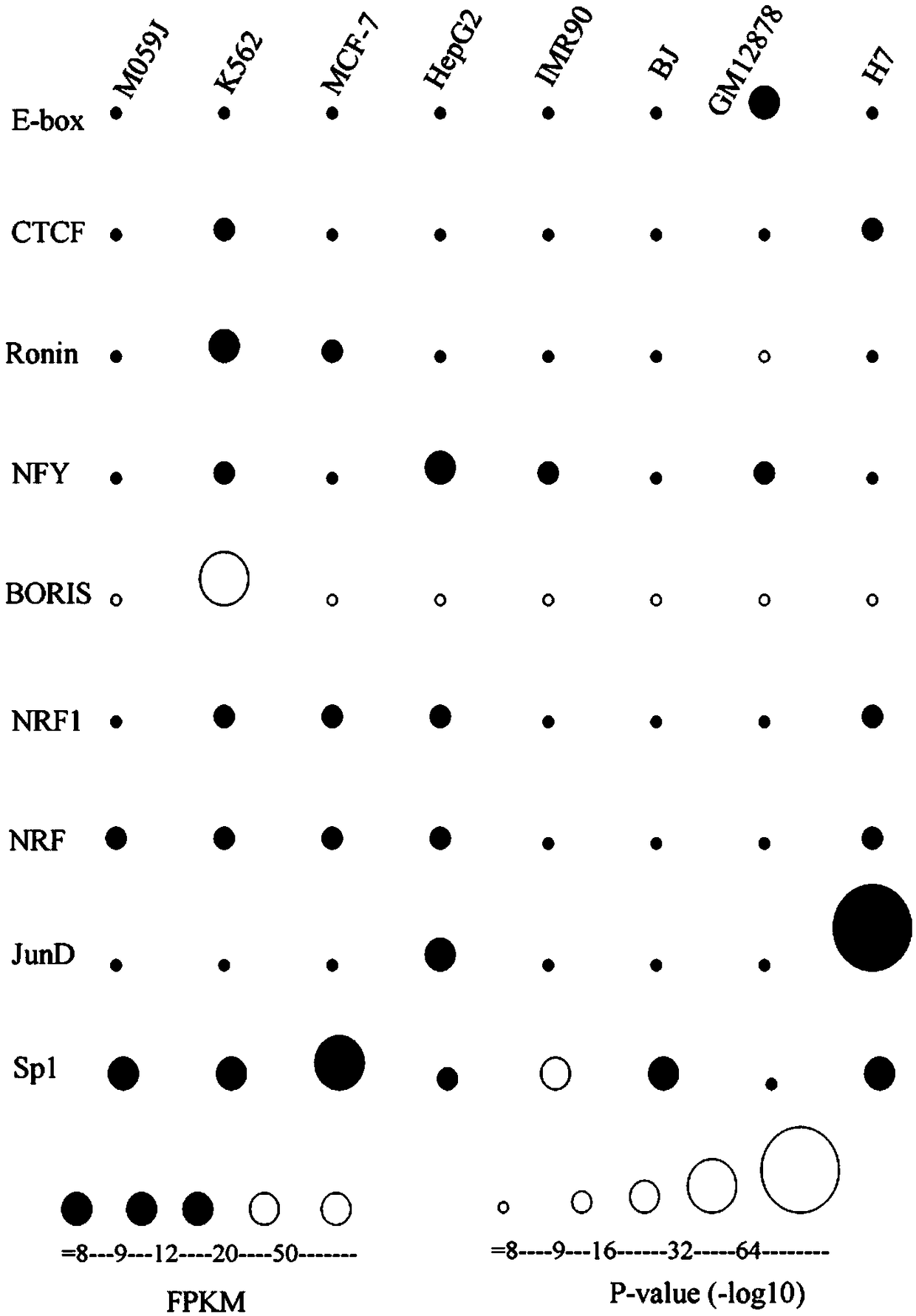
[0037] Example 2 Identifying transcription factors common to 8 cell lines and predicting the function of genomic variation in transcription factor binding sites
[0038] 1. Data source:
[0039] The high-throughput sequencing data (DNase-Seq) of 8 cell lines GM12878, IMR90, MCF-7, K562, BJ, H7, HepG2 and M059J came from the data accession number of the National Center for Biotechnology Information (GEO ID: GSE32970) (Nature 2012 Sep 6;489(7414):75-82). The cell types of the eight cell lines are listed in Table 1.
[0040] Table 1
[0041]
[0042] The sources of expression data (RNA high-throughput sequencing (RNA-Seq)) of the eight cell lines are shown in Table 1. The variation data was obtained from the genome information browser (UCSC genome browser) of the University of California, Santa Cruz, and the simple variation data information (dbSNP150) of the human genome hg19 was obtained by using the Tables function. The SNP of a single base is selected according to the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com