A method for site-directed mutation of alfalfa gene using CRISPR/Cas9 system
A gene site-directed mutation, alfalfa technology, applied in the field of gene editing, can solve the problems of incomplete RNAi silencing, limited application, unstable characteristics, etc., and achieve the effect of thorough technology, high mutation efficiency, and accelerated breeding.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
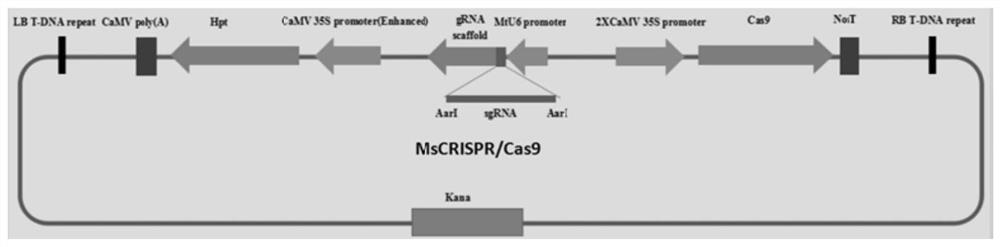
[0058] Example 1 Using the CRISPR / Cas9 system to mutate the PDS gene encoding phytoene dehydrogenase in alfalfa
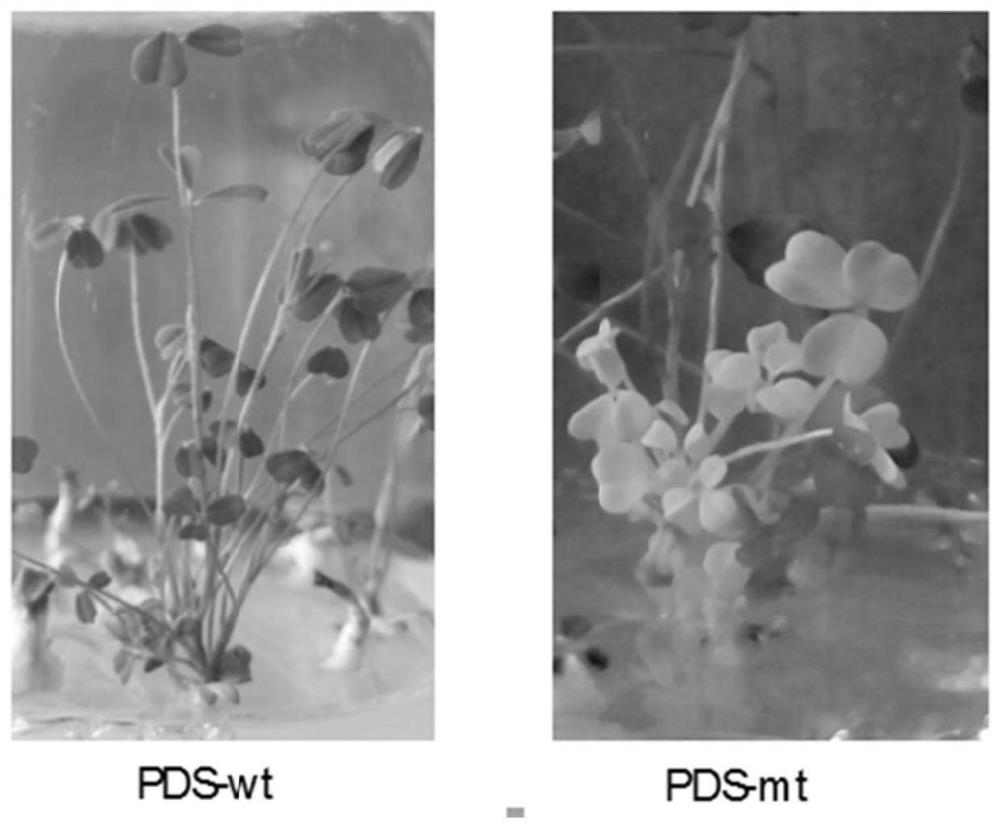
[0059] Phytoene desaturase (PDS) is the main rate-limiting enzyme in the synthetic pathway of carotenoid pigments, which can catalyze colorless C40 phytoene to produce ζ-carotene, streptosporin, tomato Red pigment, 3,4-dehydrolycopene, 3,4,3'4'-dehydrolycopene or 3,4-dehydrostreptoerythrin, etc. The mutant plants of this gene show albino phenotype, which is convenient for observation. Using this gene as a target gene in plants is a convenient way to test whether the gene knockout system works and evaluate the working efficiency.
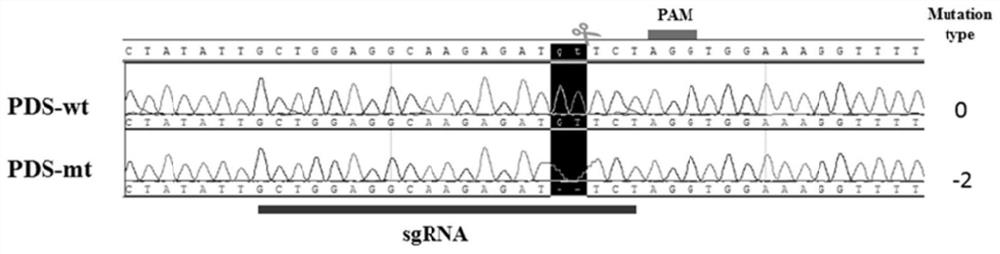
[0060] According to the PDS sequence in Medicago truncatula, a close relative of Medicago truncatula, primers were designed to amplify the PDS gene sequence of Medicago truncatula and sequenced. Based on the obtained PDS gene sequence and referring to the case in Medicago truncatula, the target site was designed; the target site was conn...
Embodiment 2
[0089] Example 2 PALM1 gene mutation in alfalfa
[0090] The difference from Example 1 is:
[0091] According to the PALM1 sequence in Medicago truncatula, a close relative of alfalfa, primers were designed to amplify the MsPALM1 gene sequence of alfalfa and sequenced, and the target site was designed according to the obtained MsPALM1 gene sequence; the target site was connected to the site of the vector MsCRISPR / Cas9 to obtain For the knockout vector MsCRISPR / Cas9::PALM1 of the MsPALM1 gene, the vector was transformed into alfalfa explants by the above-mentioned transformation method of Agrobacterium tumefaciens, and regenerated plants were obtained; the detection method was PCR-RE, and the obtained regenerated Genomic DNA samples were extracted from the plants, amplified with specific amplification primers containing the target sequence for the MsPALM1 gene, and screened for mutants by cutting PCR products with BstU I restriction endonuclease ( Figure 4 ); and by phenotypi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com