Esophageal cancer prognosis biological marker and detection method and application thereof
A biomarker and detection method technology, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of poor clinical application effect, high detection cost, time-consuming and labor-intensive detection process, etc., and achieve convenient and rapid detection , The experimental process is simple, and the effect of a wide range of applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
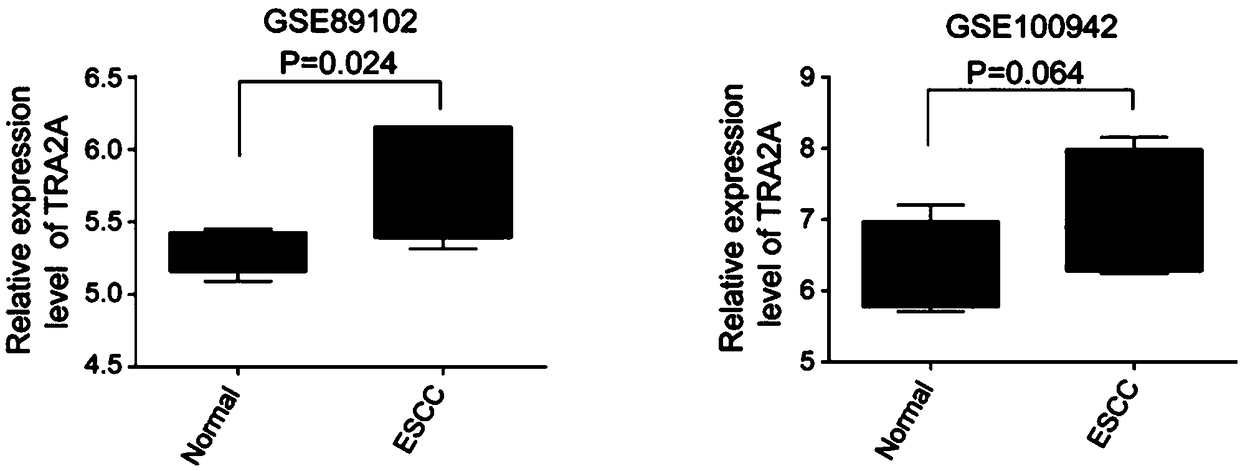
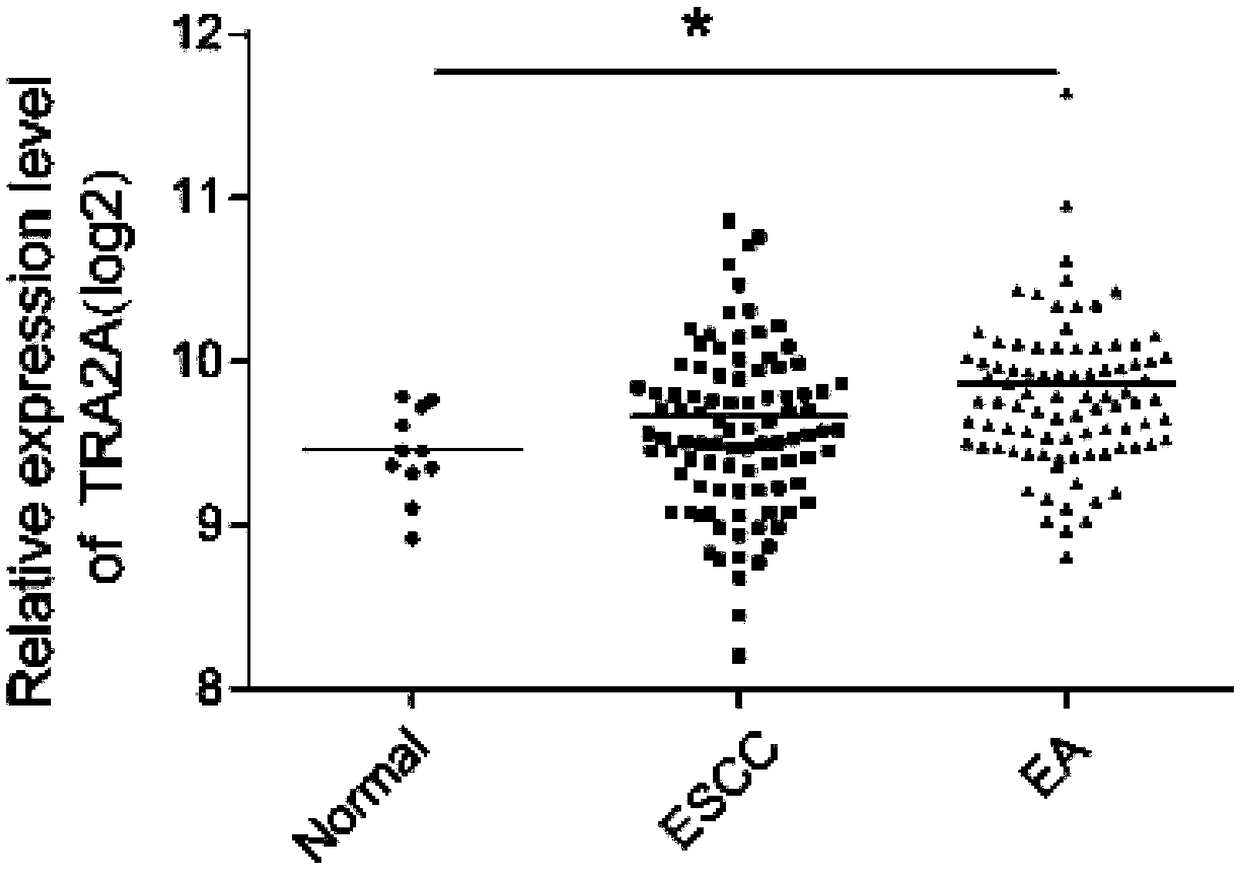
[0038] Two sets of gene expression profile data (GSE89102 and GSE100942) were downloaded from the Gene Expression Omnibus database. GSE89102 contained 5 esophageal squamous cell carcinoma patient samples and their adjacent tissue expression profile data, and GSE100942 contained 4 esophageal squamous cell carcinoma patient samples and their adjacent tissue. Tissue expression profiling data. After analyzing the two sets of expression profiles, it was found that the expression of TRA2A gene in esophageal squamous cell carcinoma tissue was significantly higher than that in adjacent tissue (GSE89102p=0.024; GSE100942, p=0.064) ( figure 1 ). At the same time, the data of 194 esophageal samples from the TCGA database were analyzed (184 cases of esophageal cancer, including 89 cases of esophageal adenocarcinoma, 95 cases of esophageal squamous cell carcinoma, and 10 cases of adjacent cancer), the same TRA2A mRNA expression was highly expressed in esophageal cancer, and in Higher expr...
Embodiment 2
[0040] Esophageal cancer patients and adjacent normal control tissues were collected, and DNA and RNA in each sample were extracted with a DNA / RNA AllPrep (Qiagen) kit. The total amount of DNA is greater than 7 μg, and the total amount of RNA is greater than 5 μg. Use RNA6000Nano assay (Agilent) to detect RIN (RNA Integrity Number) ≥ 7.0. DNA samples were detected using the Affymetrix SNP 6.0 chip platform, and the GISTIC 2.0 algorithm was used to analyze gene copy number variation analysis; RNA samples were sequenced using the IlluminaHiSeq2000PE75 platform, and the BWA algorithm was used to compare the reference genome. The number of reads in base length) represents the amount of gene expression.
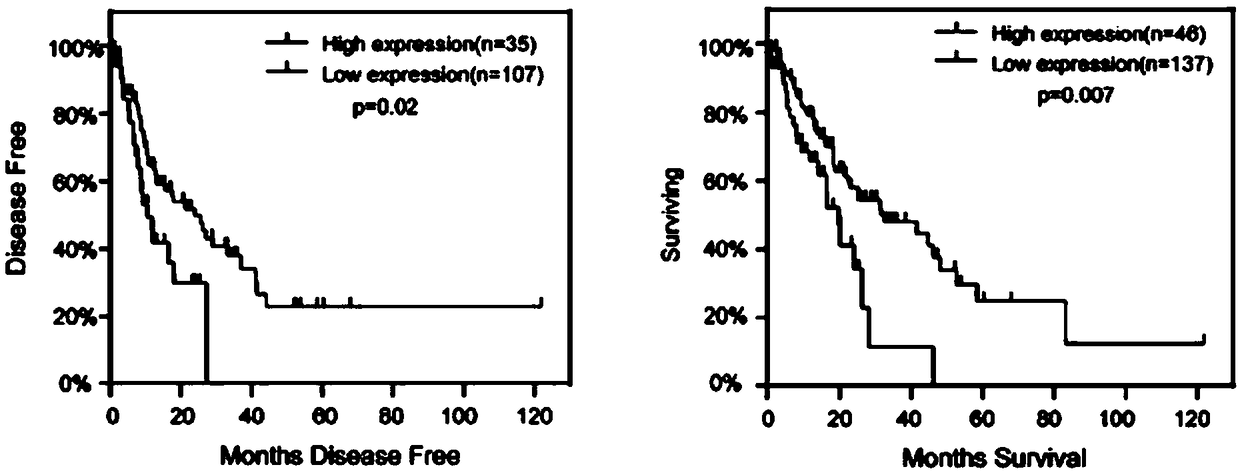
[0041]The data of 184 cases of esophageal cancer samples from TCGA were obtained according to the above-mentioned experimental method. Among them, 12% of 23 patients had TRA2A gene changes including amplification, censoring, and mRNA up-regulation. Among them, 12 cases of esoph...
Embodiment 3
[0043] Interfering with TRA2A inhibits cancer progression in esophageal cancer
[0044] In order to further confirm the role of TRA2A in the process of esophageal cancer, two siRNAs, TRA2AsiRNA#1 and TRA2A siRNA#2, were designed to interfere with TRA2A mRNA. At the same time, 48 hours after transient transfection of siRNAs in esophageal cancer cells TE1, the cells were collected, RNA was extracted by Trizol reagent, cDNA was synthesized with Takara’s PrimeScript reverse transcription kit, and Takara’s real-time quantitative kit (RR420A) was used with TRA2A-specific primers , the expression level of TRA2A gene messenger RNA was detected by Real time-PCR experiment, so as to verify the interference efficiency of siRNA.
[0045] TRA2A specific primers are:
[0046] 5' end primer CTCCAATGTCTAACCGGAGAAGA
[0047] 3' end primer TTGACACCACTCAATGGTCCA.
[0048] In TE1 cells transfected with TRA2A siRNA#1, the expression level of TRA2A mRNA was 44% compared with the control group, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com