Detection method for genomic copy number variation and device comprising same
A copy number variation and detection method technology, applied in the field of detection methods and devices including the method, can solve the problems of low accuracy, redundant steps, high false positive detection results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Example 1 Assembly of a device for detecting gene copy number abnormalities
[0084] The following modules were assembled as a means to detect genomic copy number variation:
[0085] (1) Sequencing data acquisition module: used to acquire the raw data of the sample and perform quality control and cleaning;
[0086] (2) Sequence comparison module: used to compare, sort and deduplicate the sequencing data with the reference genome;
[0087] (3) Data processing module: used to divide the reference genome into at least two windows of different sizes, and calculate the number of uniquely aligned sequences falling into the windows;
[0088] (4) GC correction module: used for counting the GC content falling into the window described in the data processing module, and performing GC correction;
[0089] (5) Reference correction module: used to use the reciprocal of the median of the window count results obtained after GC correction as a weight for reference correction, and to ...
Embodiment 2
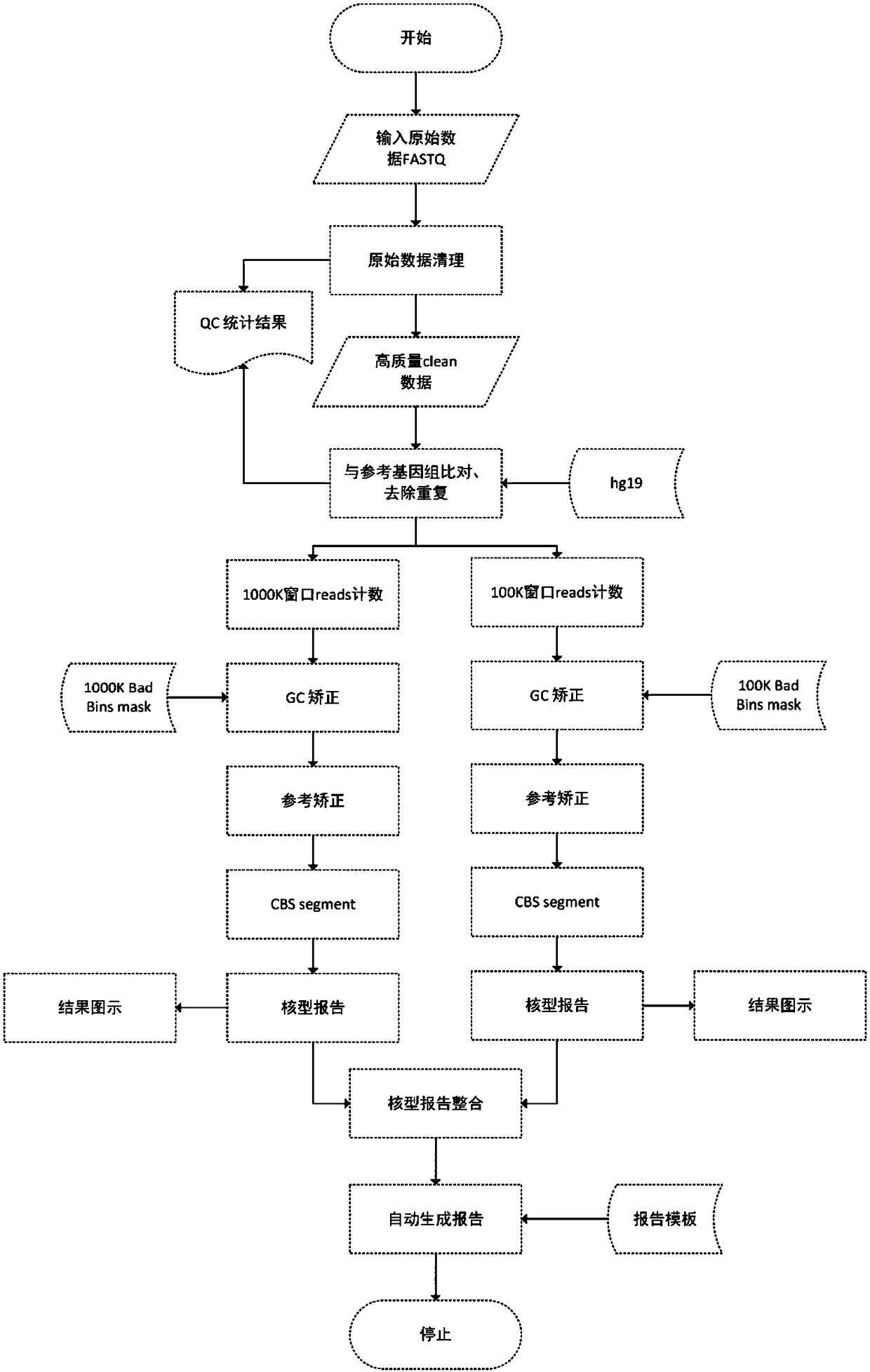
[0095] In the present invention, the device in Example 1 is used to detect copy number variation, and the complete data flow chart is shown in figure 1 As shown, the specific steps are as follows;
[0096] 1. Amplify the whole genome, build a library, and sequence the sample
[0097] In this example, the test sample is a national reference product for preimplantation chromosomal aneuploidy, which is used for performance evaluation of preimplantation chromosomal aneuploidy detection kits by high-throughput sequencing, and the evaluation is high The detection capability of a throughput sequencing method for CNVs of different chromosomal sizes in blastocyst screening.
[0098] The whole genome amplification method used the MALBAC-LAB early embryo preimplantation chromosomal aneuploid detection library preparation kit, and the amplification and library construction method was operated in accordance with the product instructions provided by Shanghai Yikang Medical Laboratory Co., ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com