Method for judging specific individual attribution of malignant tumor tissues, and application of method
A technology for malignant tumors and individuals, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of STRs mutations, SNPs detection methods are complicated to operate, and the sequencing throughput is low, achieving low mutation rate and detection The method is easy to operate and has strong scalability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] This example provides a method for judging the specific individual attribution of malignant tumor tissue: a cancer patient suspects that the hospital misadjusted the tumor tissue wax block after surgery to remove the tumor, resulting in a misdiagnosis, and requests to identify whether the archived tumor tissue wax block comes from himself Include the following steps:
[0035] 1. Reference sample
[0036] Take 0.5ml of the patient's peripheral blood to make a blood card as a reference sample, and dry it for later use.
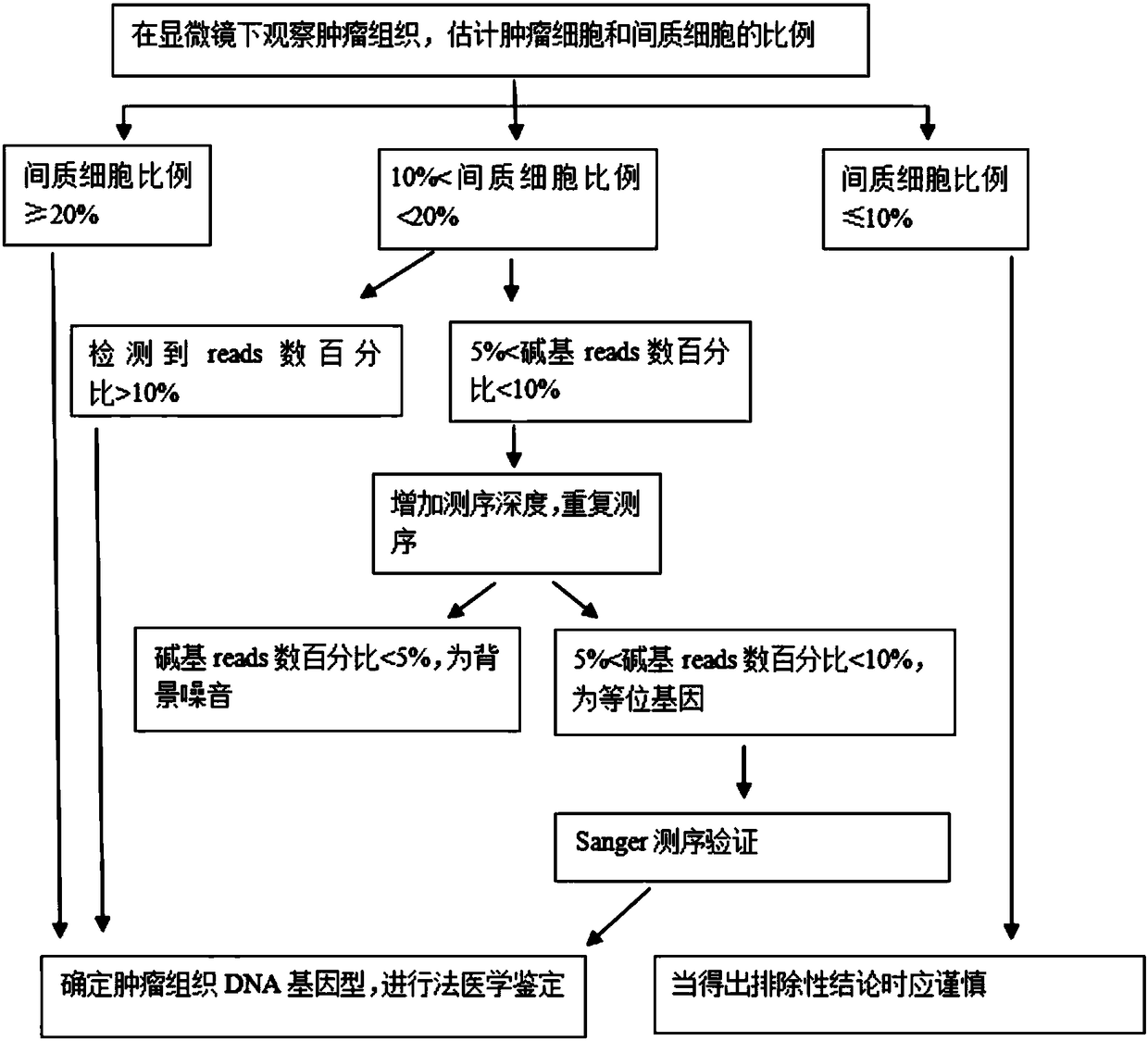
[0037] 2. Process tumor tissue samples to determine the percentage of stromal cells
[0038] Make 5 μm thick sections with tumor tissue wax blocks, stain with hematoxylin and ethanol (HE), and observe the sections under a microscope. First use 40× magnification to find the most infiltrated area of tumor tissue, and then use 100× magnification to evaluate the tumor The percentage of the area occupied by the cells in the visual field, and the rest is the ...
Embodiment 2
[0054] This example provides a method for judging the specific individual attribution of malignant tumor tissue: a cancer patient suspects that the hospital misadjusted the tumor tissue wax block after surgery to remove the tumor, resulting in a misdiagnosis, and requests to identify whether the archived tumor tissue wax block comes from himself Include the following steps:
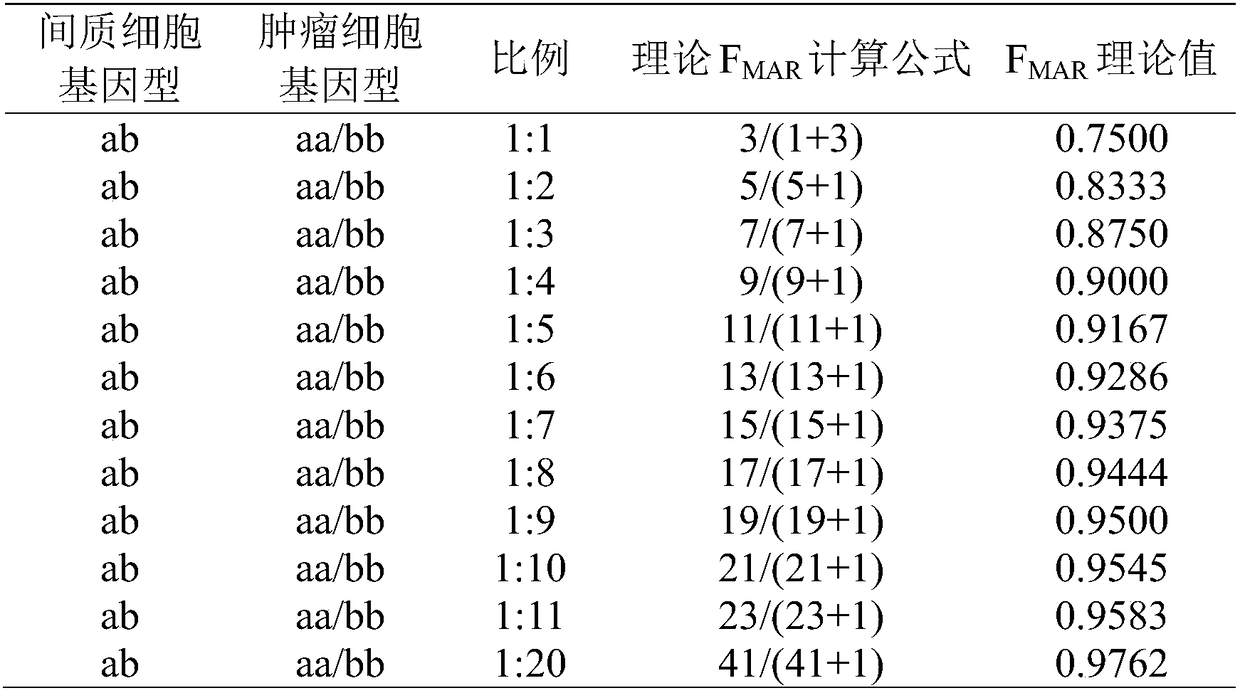
[0055] Steps 1 to 6 are the same as in Example 1, wherein the ratio of tumor tissue-to-stromal cells is 30%.
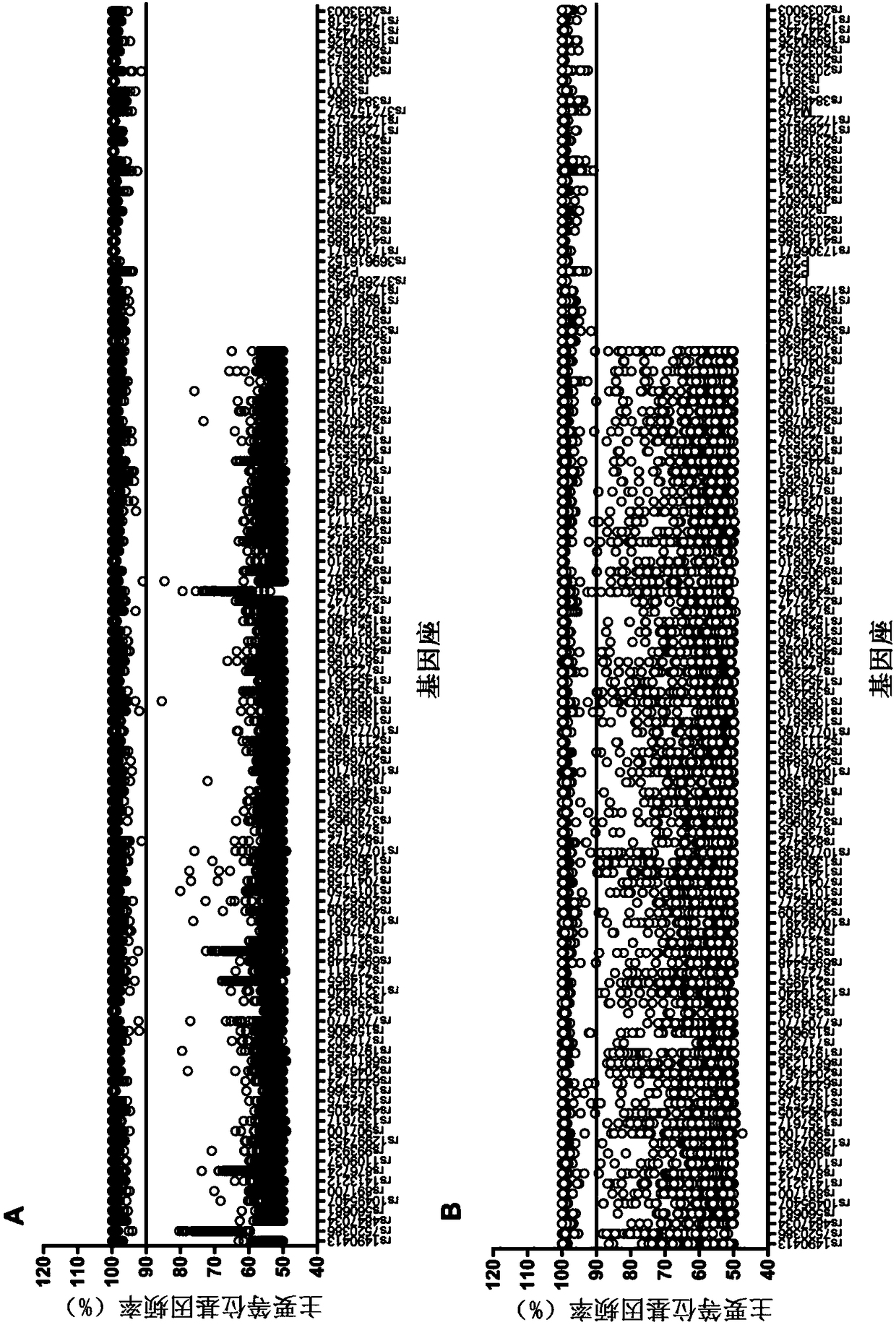
[0056] 7. Check the SNP typing of tumor tissue and peripheral blood samples of patients. (1) "NN" appears in the typing result, indicating that the plug-in cannot judge the type after comprehensive detection of the sequencing data. Download the BAM and BAI files generated after the sequencing data analysis, and import them into the IGV software for visual analysis. According to the principle of min_snp_coverage≥50 and min_allele_frequency>10%, the site is manually typed; (2) All heterozygous loci...
Embodiment 3
[0059] This example provides a method for judging the specific individual attribution of malignant tumor tissue: a cancer patient suspects that the hospital misadjusted the tumor tissue wax block after surgery to remove the tumor, resulting in a misdiagnosis, and requests to identify whether the archived tumor tissue wax block comes from himself Include the following steps:
[0060] Steps 1 to 6 are the same as in Example 1, wherein the ratio of tumor tissue to mesenchymal cells is 10% to 20%.
[0061] 7. Check the SNP classification of tumor tissue, (1) "NN" appears in the typing result, indicating that the plug-in cannot judge the type after comprehensive detection of the sequencing data. Download the BAM and BAI files generated after the sequencing data analysis, and import them into the IGV software for visual analysis. According to the principle of min_snp_coverage ≥ 50 and min_allele_frequency > 10%, the site is manually typed; (2) in tumor tissue classification In the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com