Universal primer single fluorescence molecule detection technique and kit
A universal primer and molecular detection technology, applied in the field of nucleic acid detection, can solve problems such as false positives and interference of experimental results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0130] Embodiment 1, the establishment of the nucleic acid detection method based on the multiple digital PCR of universal primer
[0131] In this example, transgenic corn was used as a sample, four transgenic screening elements, P-35S, T-NOS, NPTII, and PAT, and the internal reference adh1 of corn were selected as the target gene, and a multiplex nucleic acid detection method based on digital PCR was established.
[0132] 1. Grinding of the sample to be tested
[0133] (1) Dry all non-transgenic negative samples and transgenic samples in hot air at a lower temperature to reduce the moisture content in crop seeds and protect the genome from degradation;
[0134] (2) Put the dried sample into a ball mill for pulverization, the pulverization procedure is pulverization for 50 seconds, and place it at room temperature for 1 min, and lower the pulverized sample to room temperature to reduce the degradation of the genome during the pulverization process;
[0135] (3) Collect the po...
Embodiment 2
[0162]Example 2, Optimization of the nucleic acid detection method of multiplex digital PCR based on universal primers
[0163] (1) Optimization of universal primers and probes
[0164] 1. Design of primers
[0165] The universal primer modifies a common sequence with the same nucleotide sequence at the 5' end of each pair of primers, and any specific primer modified at the 5' end by the universal primer is called a modified specific primer. When there are modified specific primers and universal primers in the reaction system at the same time, in the first stage of the first ten cycles of the PCR process, the modified specific primers play a role in amplifying the target sequence with universal primers at both ends, and the universal primers play a huge role in the second stage. To ensure that each target sequence has a uniform amplification efficiency. Universal primers can effectively reduce the interference between multiple pairs of primers, so that multiple pairs of prim...
Embodiment 3
[0222] Embodiment 3, the sensitivity experiment of the nucleic acid detection method based on the multiplex digital PCR of universal primer
[0223] (1) Absolute limit of quantitation aLOD and absolute limit of detection aLOQ of the method
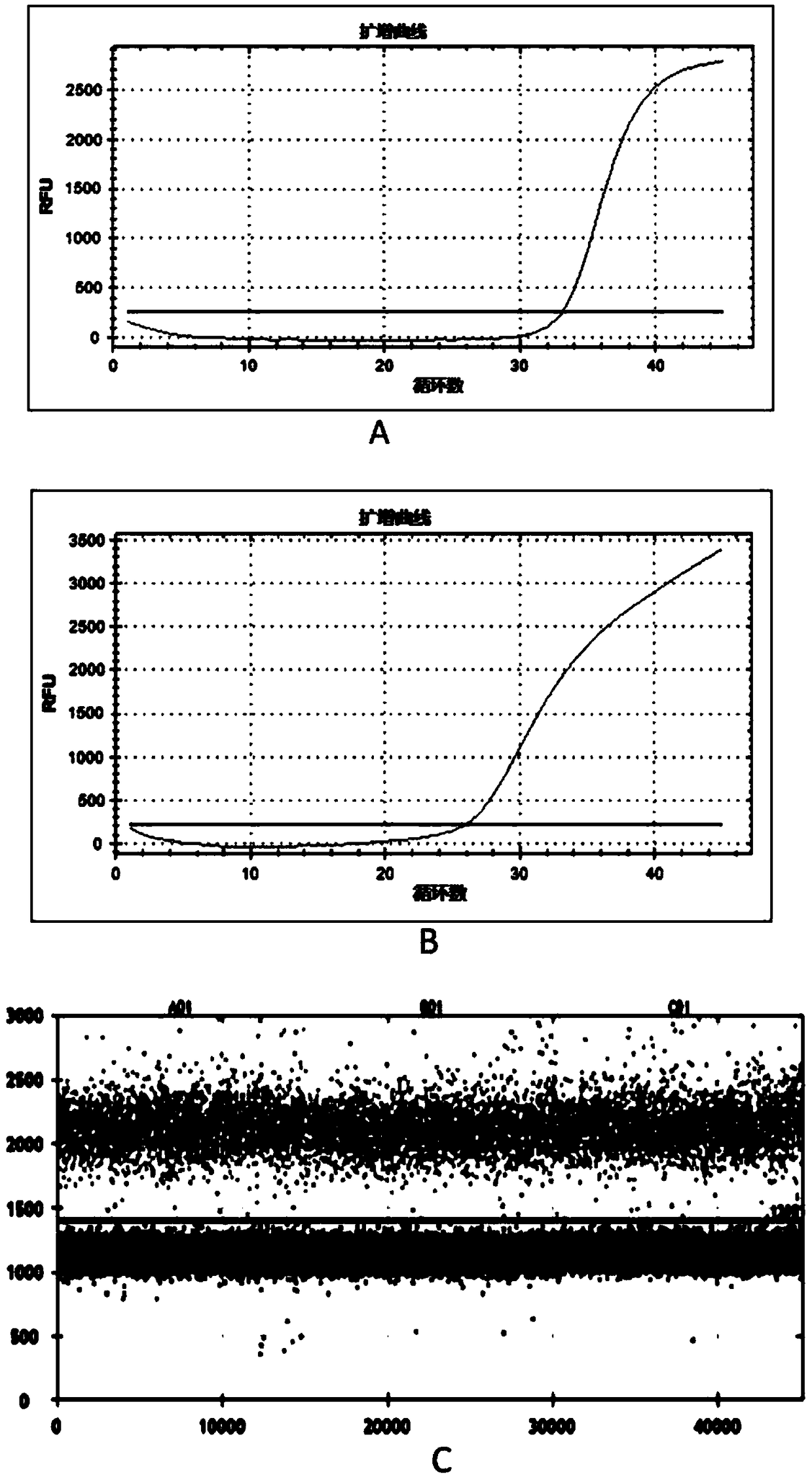
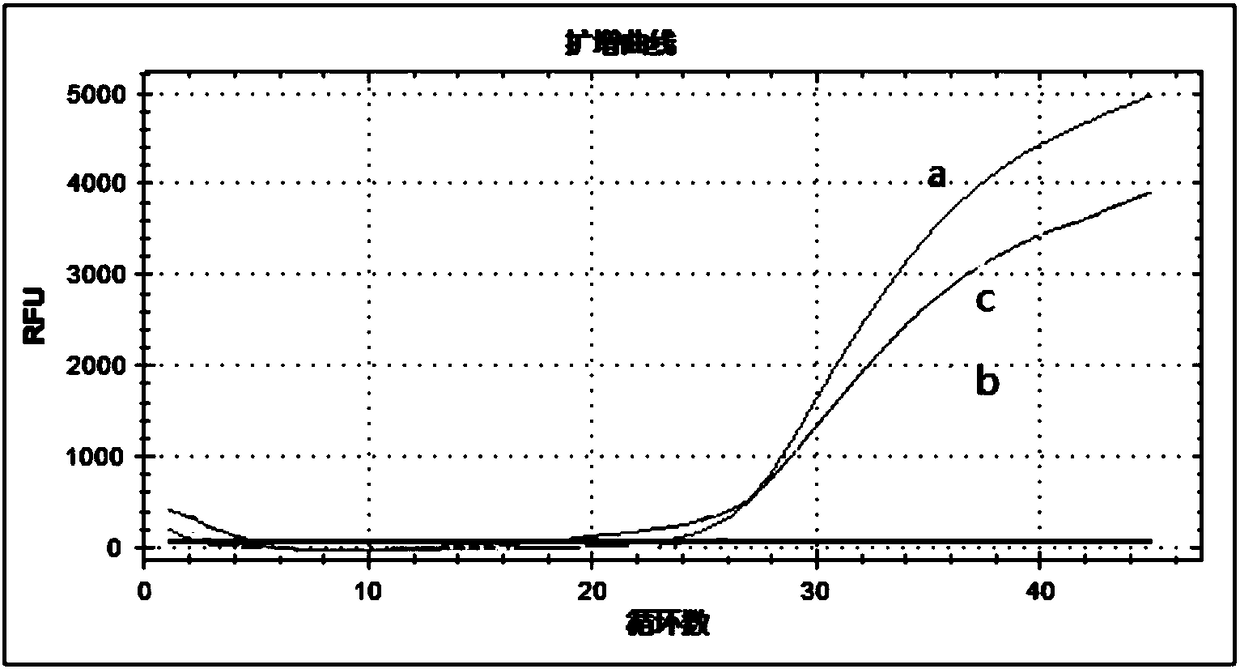
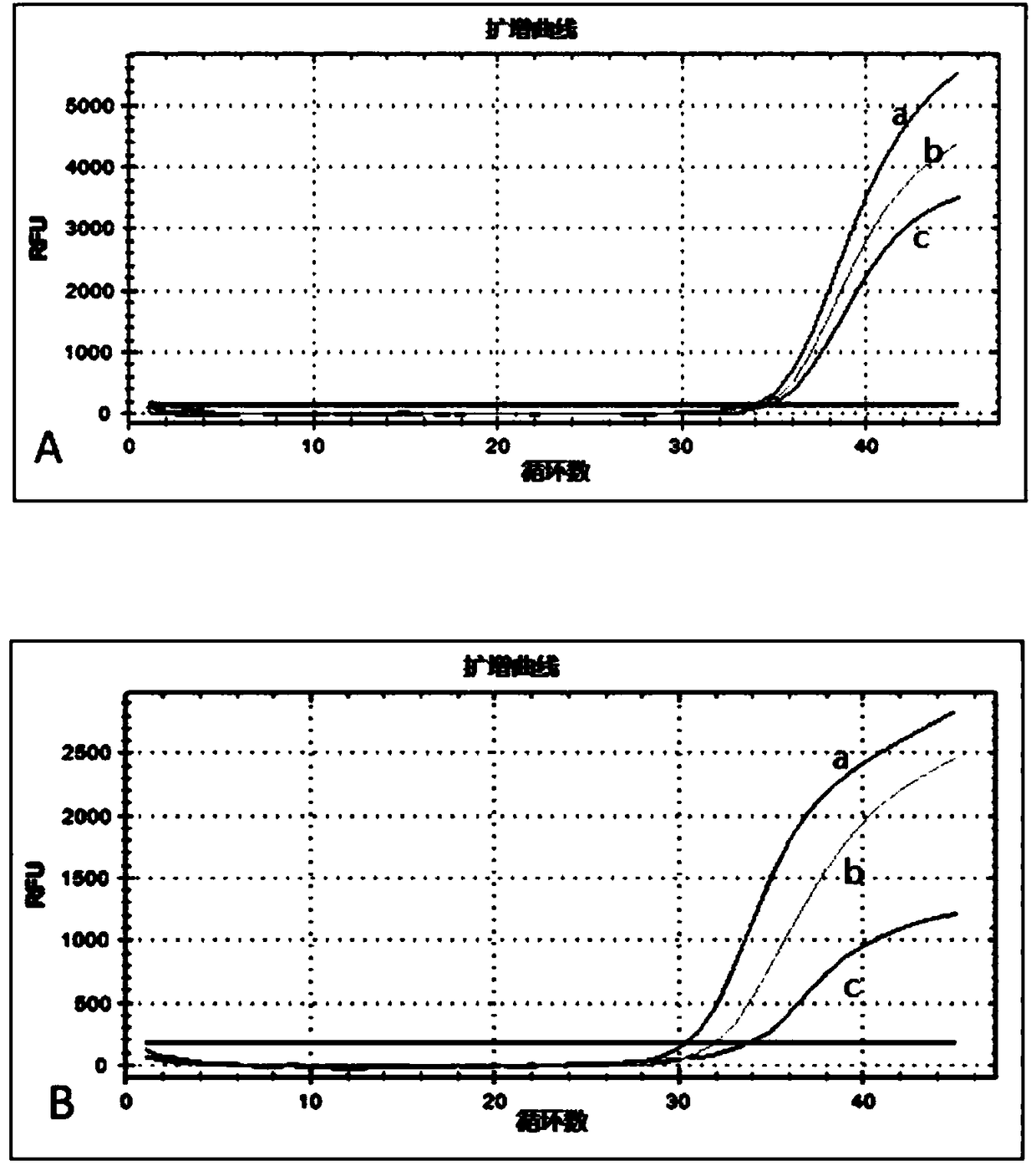
[0224] In order to explore the detection limit of this method, the genomes of transgenic maize MON863 and BT11 (gifted by Monsanto) (50ng / μl, solvent is water) and BT11 (50ng / μl, solvent is water) were uniformly mixed at a volume ratio of 1:1 , and then dilute the mixture with water to prepare a series of samples to be tested, which are recorded as 5x, 10x, 50x, 100x, 500x, 1000x, 5000x and 10000x respectively.
[0225] Carry out the sensitivity test according to the multiplex digital PCR amplification method described in Step 4 of Example 1, the difference is that the above-mentioned diluted series of samples to be tested are respectively replaced successively with the genomic DNA components in the reaction system shown in Table 2, the re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com