Preparation method of nucleic acid targeted capture sequencing library based on long chain molecule inversion probe
A molecular inversion probe and sequencing library technology, which is used in the field of nucleic acid target sequence capture sequencing library preparation based on primer extension, and can solve problems such as limited MIP or Padlock probe application, quantification of DNA fragments that cannot be captured, and limited capture length. , to achieve the effect of wide application range, convenient operation and improved capture efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0060] The present invention will be described in further detail below in conjunction with the accompanying drawings and specific embodiments.
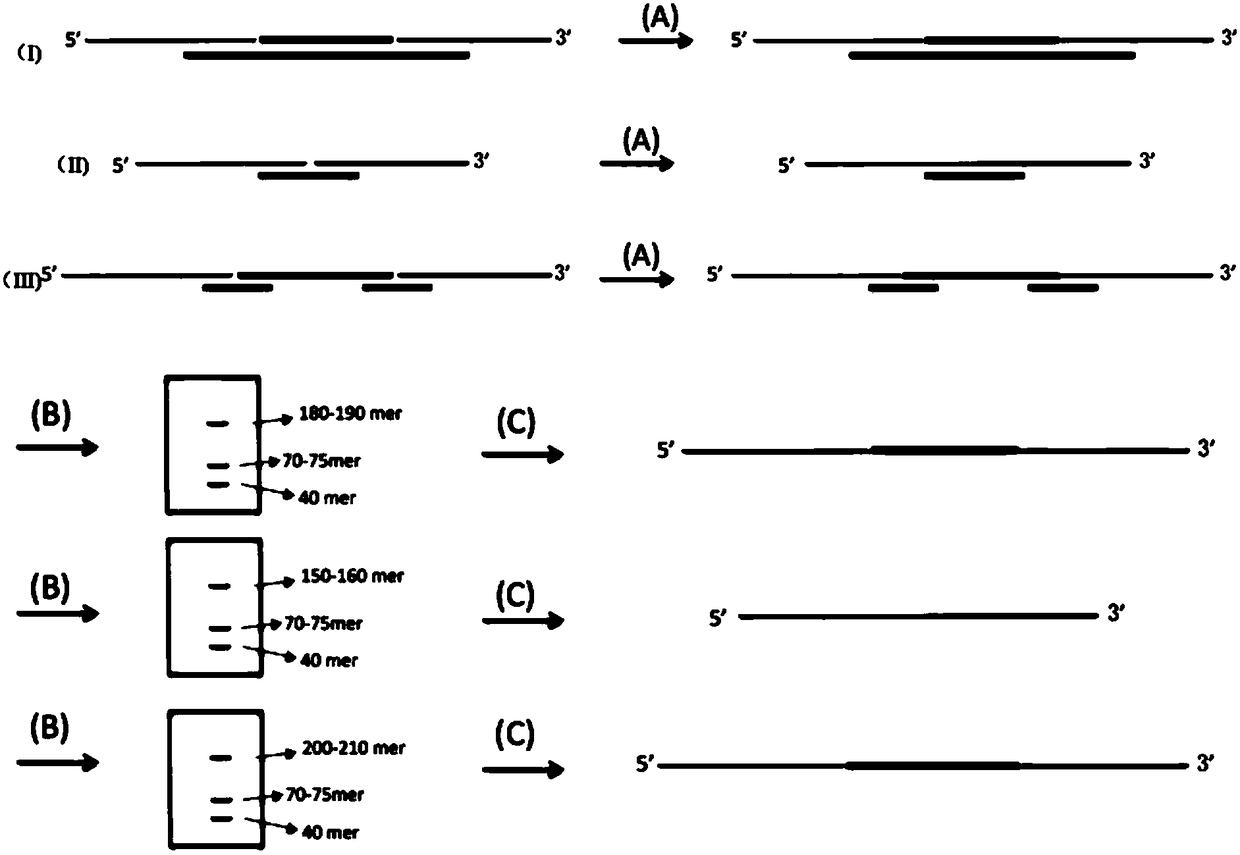
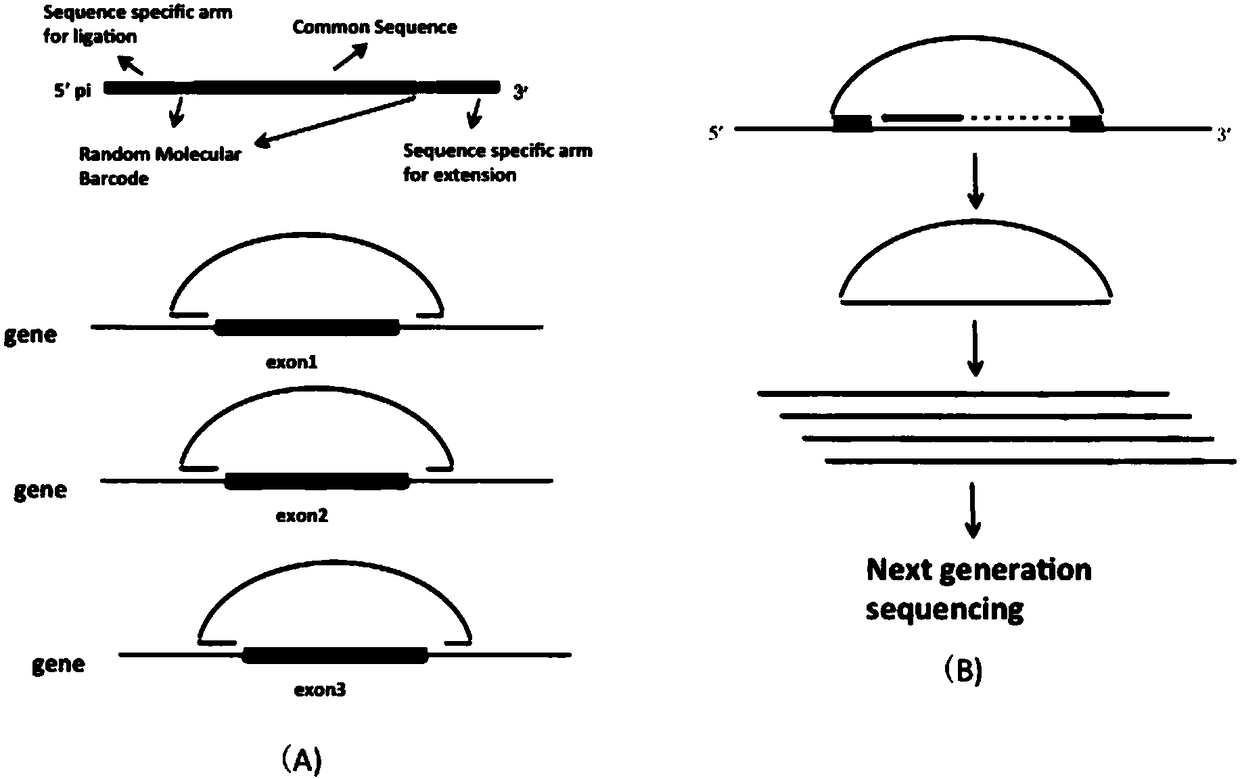
[0061] figure 1 Shown is the preparation method of the long-chain MIP probe of the present invention, and there are three specific preparation processes. Such as figure 2 Shown are the schematic structure of the prepared long-chain MIP probe (top panel of A) and the capture design diagram of the target region (eg, exon) (3 panels below A). A nucleic acid targeted capture sequencing library preparation method based on long-chain molecular inversion probes, the method comprising the following steps: 1) long-chain probe preparation process
[0062] (a) The first long-chain probe preparation process: the process is as follows figure 1 As shown in (I), design and synthesize the oligo sequences of probes A1-A50 and B1-B50 and the public sequence a sequence (pi-acaaaggtaagtcaagtgactcttgatgtttgtctcatca) and the b sequence reversely comple...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com