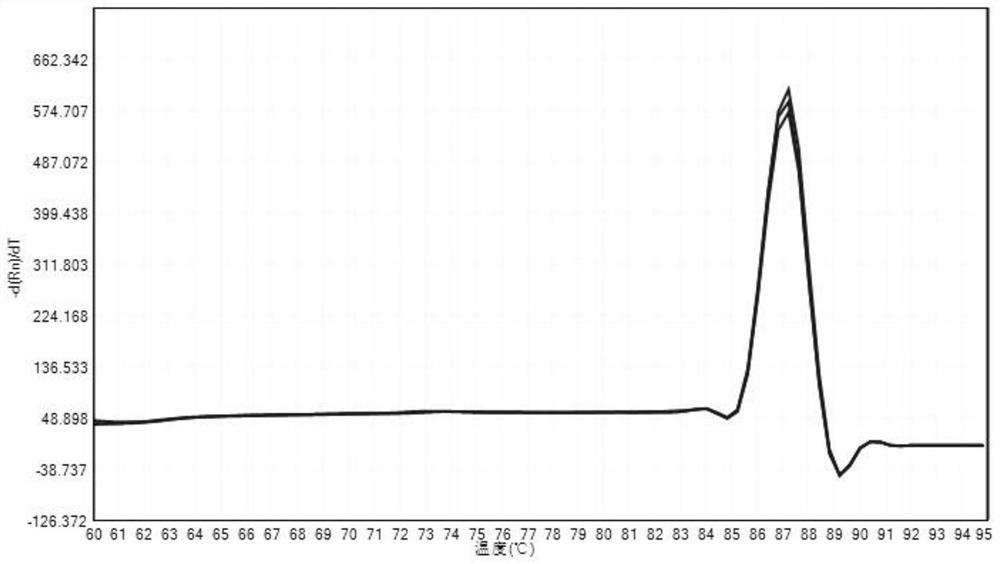
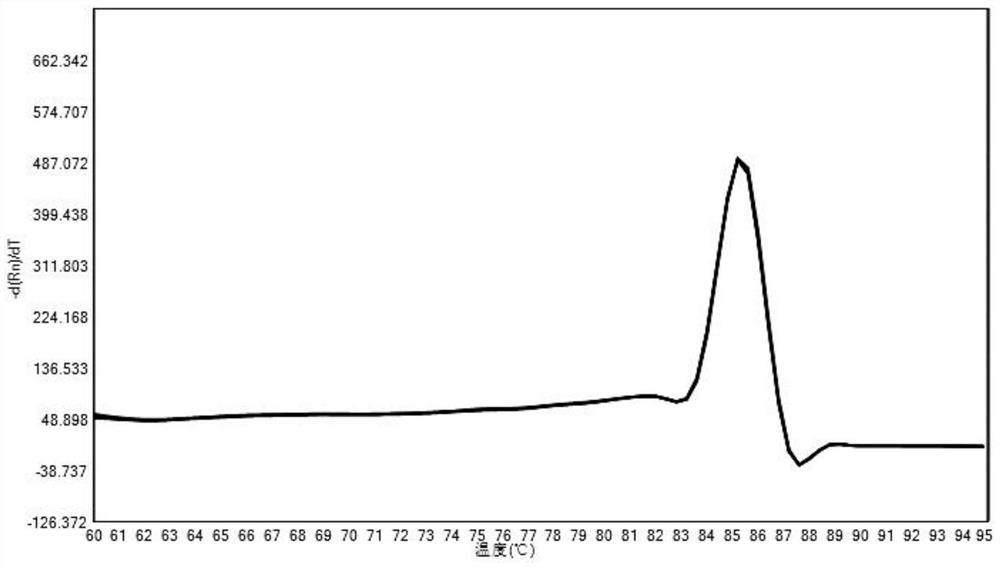
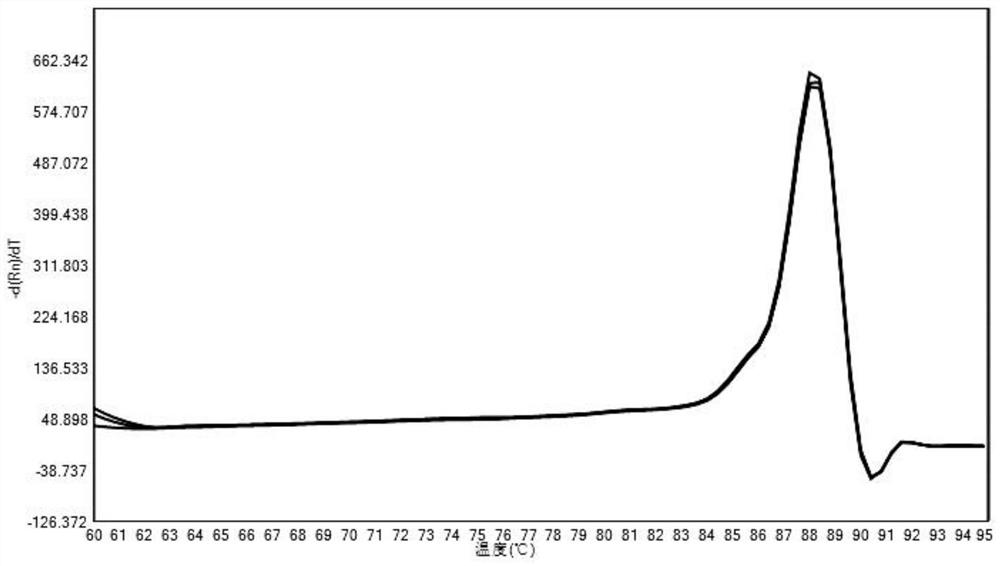
A method for detecting the relative content of bacterial species in a sample
A technology for relative content and detection of samples, applied in the biological field, can solve cumbersome problems and achieve the effect of overcoming cumbersome operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1 This embodiment discloses a method for determining the relative content of Bifidobacterium and Lactobacillus in the total number of bacteria in a feces sample. The specific steps are as follows:
[0053] 1 sample pretreatment
[0054] 1.1 Spread a layer of plastic wrap on the surface of the ultra-clean workbench, and prepare weighing paper, disposable blades, and small spoons.
[0055] 1.2 Put on disposable PE gloves and take a half-thawed sample. Place the half-thawed sample on weighing paper, and cut off about 200 mg of the inner part (the part not exposed to the air) with a disposable blade before it is completely thawed, and put the cut part into a centrifuge tube with a blade or a spoon , complete the sampling.
[0056] 1.3 The sample after sampling is stored at -80°C. Throw the used weighing paper into the trash can, replace the disposable blade, put the blade into the sharps box, wipe and clean the sampling spoon with medical alcohol.
[0057] 1....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com