Simplified representative unicellular whole-genome database creating method and application thereof
A whole-genome, single-cell technology, applied in biology and simplified genome field, which can solve problems such as base errors, SNV false positives, single nucleotide variations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0126] Simplified Genomic Library Construction of Human Peripheral Blood Samples
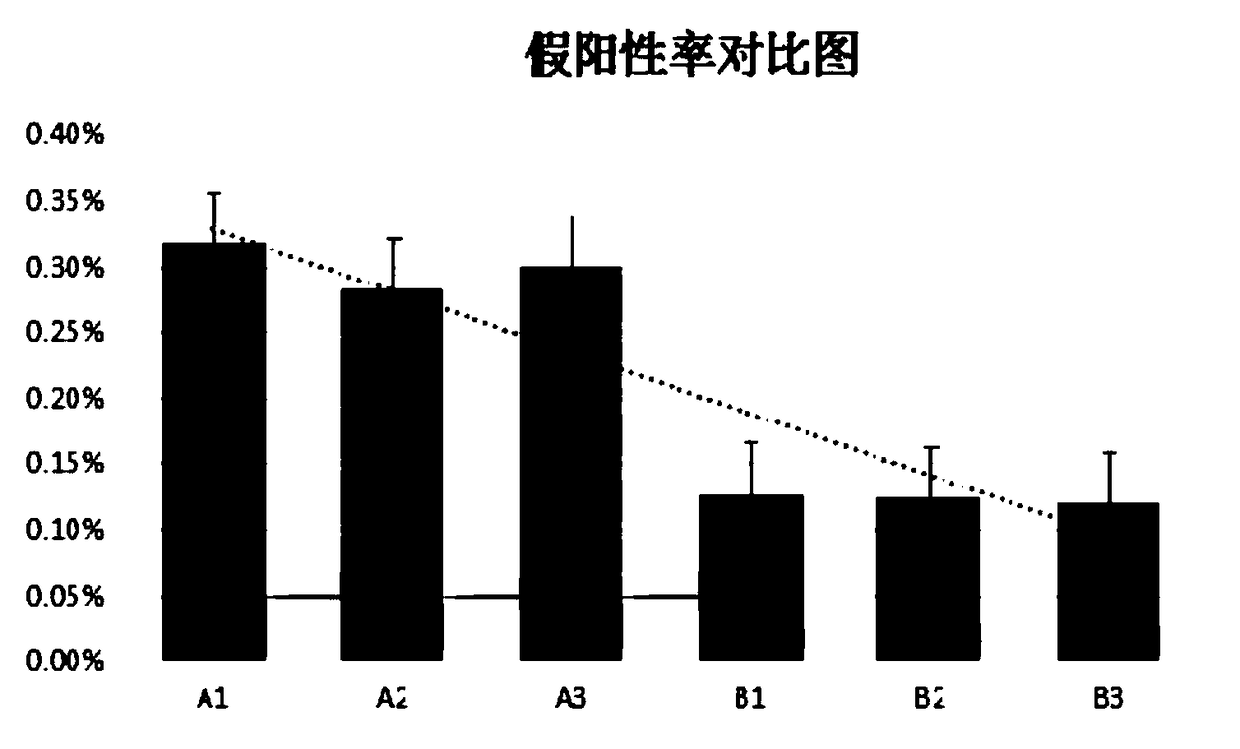
[0127] 1. Take a peripheral blood sample and divide the separated white blood cells into 6 tubes, with 1 cell in each tube to ensure that the same human single cell sample is obtained. Divided into two groups, A and B, respectively named A1 / A2 / A3, B1 / B2 / B3. Group A was amplified according to the above-mentioned technical scheme except that the enzyme treatment step in step 2 was not added, and group B was amplified and library-built completely according to the above-mentioned technical scheme. Take 500 microliters of blood from the same tube for whole blood DNA extraction, named as bulk, for subsequent false positive comparison.
[0128] 2. After the above-mentioned amplified product is purified by magnetic beads, the sequencing depth is 100 times. The bulk samples were simultaneously constructed and sequenced.
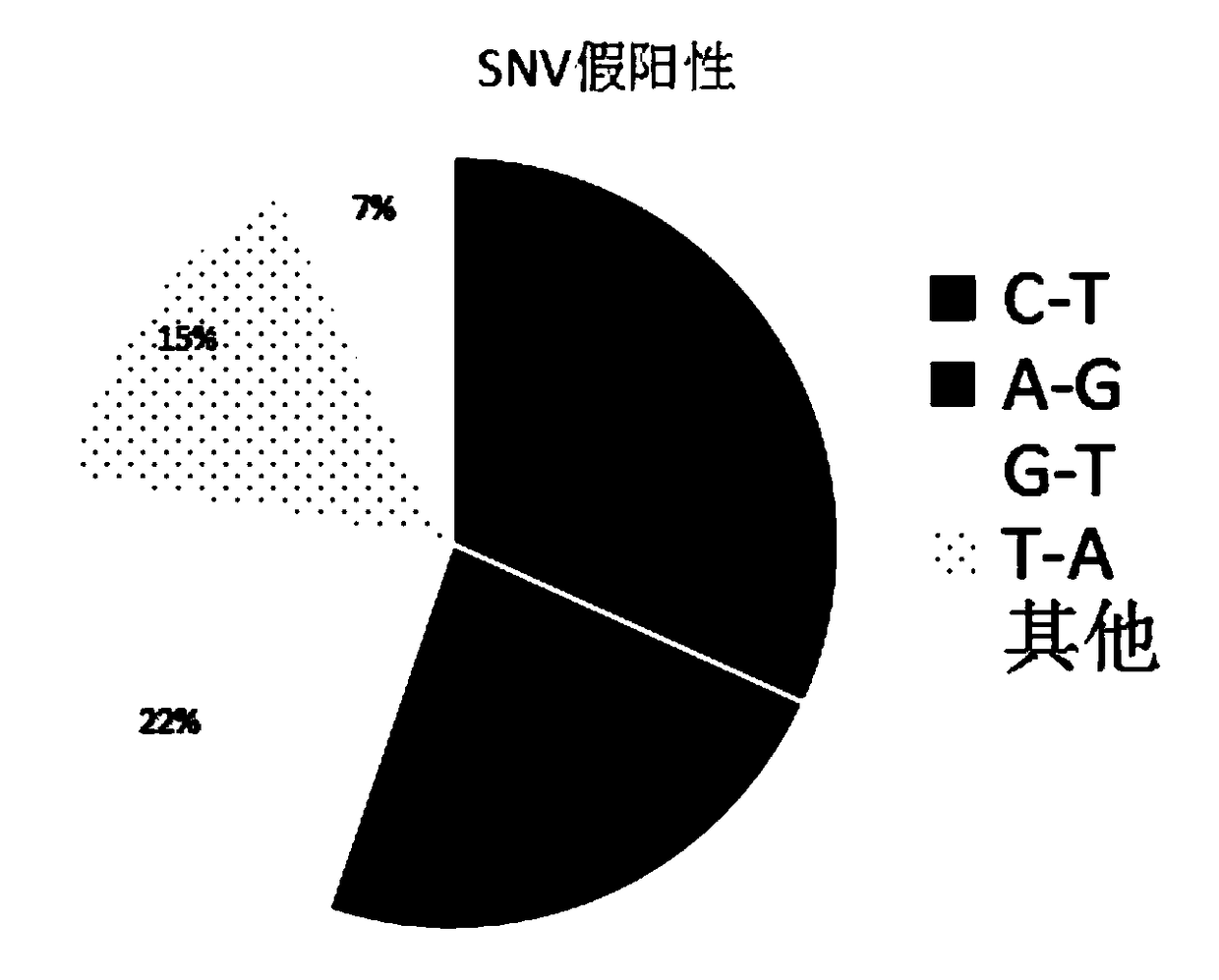
[0129] 3. Data analysis: Statistics of the base mutation types and ratios betwee...
Embodiment 2
[0143] To study the genomic differences between catfish single fertilized egg samples in two different regions (region A, region B), the amplification of a single fertilized egg sample and library construction were carried out according to the process in the technical plan, and a single fertilized egg sample of area C perch was used to carry out Conventional genomic library construction, as a control analysis sample. The results of the association analysis are as follows. For the FST analysis of the distribution of the genetic parameters of each population on the genome, please refer to Figure 4 As shown, for the PCA analysis on it, please refer to Figure 5 shown.
[0144] according to Figure 4 , 5 The specific analysis is as follows: FST means F-statistics (F analysis) in genetic analysis. Fst is a measure of genetic differentiation between subpopulations. The extent of genetic relationships between different populations can be quantified. FST analysis found that alt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com