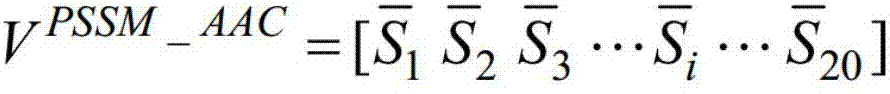Method for predicting outer membrane protein of germs on basis of machine learning technique
A technique of technology prediction and machine learning, applied in proteomics, genomics, instrumentation, etc., to achieve the effect of accelerating the identification process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] The present invention is achieved in this way, a method for predicting bacterial outer membrane proteins based on machine learning technology, the method for predicting outer membrane proteins at the level of the whole genome of bacteria is:
[0040] Using the PSI-BLAST algorithm, the protein sequence is compared with the non-redundant protein sequence database, and the position-specific iterative scoring matrix (PSSM) is calculated, and the composition characteristics (amino acid residues) of the PSSM are calculated by the composition function and the autocorrelation function. Composition / PSSM_AAC), and PSSM autocorrelation features (autocorrelated amino acid position-specific composition / PSSM_AC), establish a classifier based on support vector machine, classify outer membrane proteins and non-outer membrane proteins, through local computer program, accept user The input protein sequence predicts whether the user-entered protein sequence is an outer membrane protein.
Embodiment 2
[0042] A method for predicting bacterial outer membrane proteins based on machine learning technology, the method for predicting outer membrane proteins at the bacterial genome level is:
[0043] Step 1. The user inputs the protein sequence to be predicted into the local computer program in FASTA format;
[0044] Step 2, the computer program uses the PSI-BLAST program to compare the protein sequence with the non-redundant protein sequence;
[0045] Step 3, the computer program calls Matlab to run the core prediction program, and calculates the PSSM composition characteristics and PSSM autocorrelation characteristics of the protein;
[0046] Step 4, the Matlab program performs feature selection and combination of multiple types of features according to a preset method to generate a protein feature vector;
[0047] Step 5, the Matlab program calls the libSVM program, and uses the pre-trained model to predict the likelihood that the protein is an outer membrane protein;
[0048...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com