Method and application of rapid detection of nucleic acid based on electrochemical potential pretreatment technology
An electrochemical and pretreatment technology, applied in the direction of biochemical equipment and methods, electrochemical variables of materials, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1. DNA-DNA hybrid system
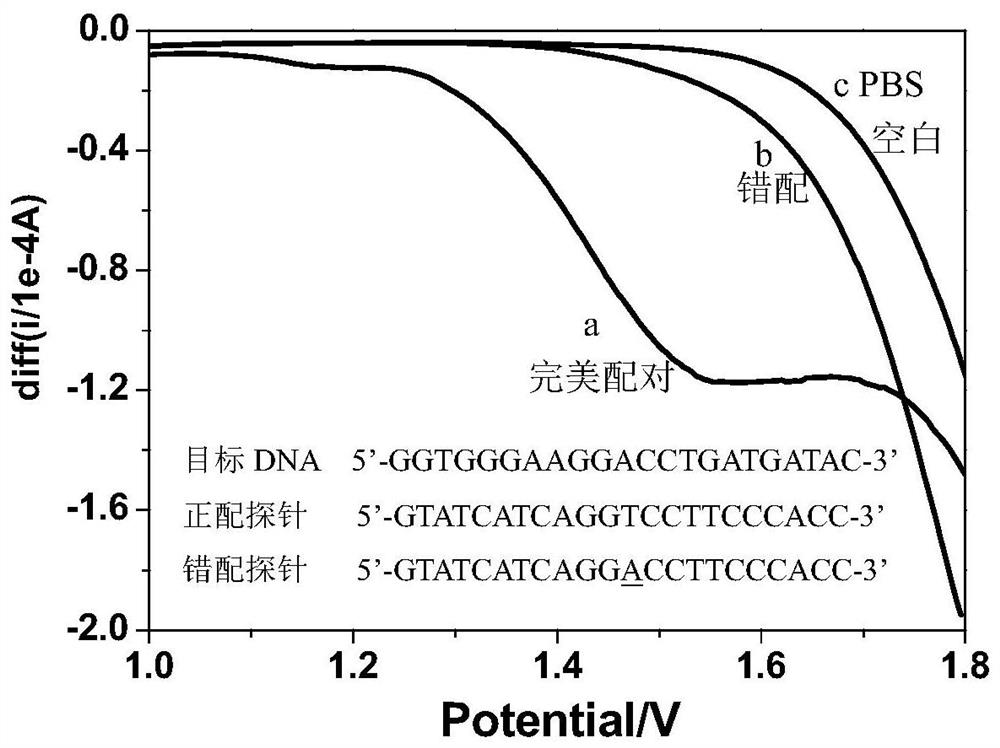
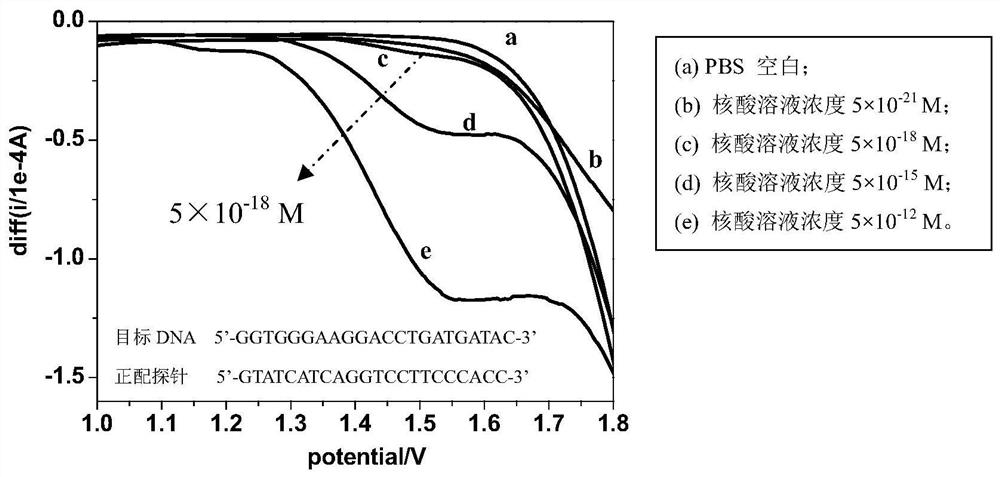
[0022] The artificially synthesized target DNA (5'-GGTGGGAAGGACCTGATGATAC-3') solution was dropped into 5pM positive matching probe (5'-GTATCATCAGGTCCTTCCCACC-3') solution and 5 pM mismatching probe (5'-GTATCATCAGG) solution respectively. A In the CCTTTCCCACC-3') solution, a positive hybridization solution and a mismatch hybridization solution are formed through the DNA-DNA hybridization reaction. The glassy carbon electrode was pretreated in the potential range of 1.8-2.7V, and then the simulated body fluid phosphate buffer solution (PBS buffer) with pH=7.4 was used as a blank comparison. The hybridization solution and the PBS blank solution were subjected to square wave voltammetry scanning, and the electrochemical sensing detection results were as follows figure 1 Shown: the square wave voltammetry curve of the positive hybridization solution has a shoulder peak in the range of 1.0-1.8V, and the square wave voltammetry curve of the...
Embodiment 2
[0023] Example 2. DNA-cDNA hybridization system
[0024] The cDNA (5'-TAACACTGTCTGGTAAAGATGG-3') solution of artificially synthesized prostate cancer characteristic marker miR-141 was dropped into 5pM positive matching probe (5'-CCATCTTTACCAGACAGTGTTA-3') solution and 5 pM mismatching probe solution respectively. Needle (5'- A In the CATCTTTACCAGACAGGTGTTA-3') solution, a positive hybridization solution and a mismatch hybridization solution are formed through the cDNA-DNA hybridization reaction. The glassy carbon electrode was pretreated in the potential range of 1.8-2.5V, and then the simulated body fluid phosphate buffer solution (PBS buffer) with pH=7.4 was used as a blank comparison. The hybridization solution and the PBS blank solution were scanned by square wave voltammetry, and the electrochemical sensing detection results were similar to those in the appendix. figure 1 , figure 2 shown.
Embodiment 3
[0025] Example 3. DNA-RNA hybrid system
[0026] The artificially synthesized target RNA (5'-GGUGGGAAGGACCUGAUGAUAC-3') solution was dropped into 5pM positive matching probe (5'-GTATCATCAGGTCCTTCCCACC-3') solution and 5 pM mismatching probe (5'-GT) solution respectively. T In TCATCAGGTCCTTCCCACC-3') solution, a positive hybridization solution and a mismatch hybridization solution are formed through DNA-RNA hybridization reaction. The glassy carbon electrode was pretreated in the potential range of 0-2.7V, and then the simulated body fluid phosphate buffer solution (PBS buffer) with pH=7.4 was used as a blank comparison. The hybridization solution and the PBS blank solution were scanned by square wave voltammetry, and the electrochemical sensing detection results were similar to those in the appendix. figure 1 , figure 2 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com