Multiple lung cancer-related gene methylation combined detection kit, multiple lung cancer-related gene methylation combined detection method, and applications of multiple lung cancer-related gene methylation combined detection kit
A detection reagent and joint detection technology, which is applied in the field of multiple lung cancer-related gene methylation joint detection kits, can solve the problems of difficult early detection and early qualitative diagnosis, and difficult early diagnosis of lung cancer, so as to improve the detection rate and design Ingenious, simple structure effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Embodiment 1, the selection of detection target gene
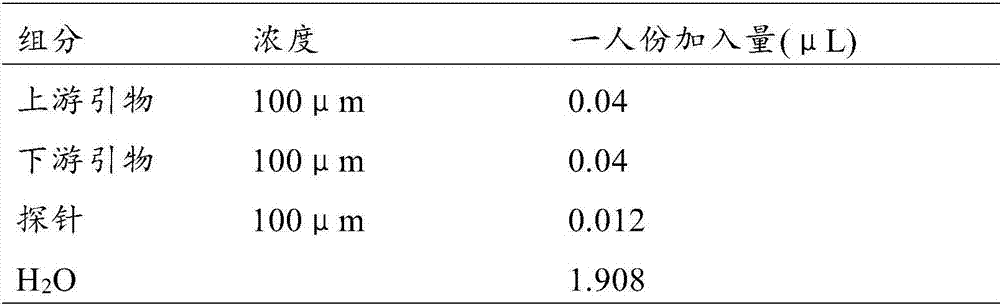
[0067] Methylated DNA has obvious advantages as a detection target. Compared with protein markers, DNA can be amplified and easily detected; compared with mutation markers, methylated DNA is located in a specific part of the gene , generally in the promoter region, making detection easier and more convenient. In the previous study, the methylated DNA detection results of different genes had a certain synergistic effect. In order to further improve the clinical positive detection rate, on the original basis (p16, H-cadherin (CDH13), SHOX2, HOXA9, RARβ, RASSF1A) were added The new ANKRD18B and MPDZ genes are used as candidate detection genes to further study the distribution of methylation sites of each gene, and design primers and probes for detection respectively. The detection primers and probes for each gene are as follows:
[0068] The detection primers and probes for p16 are:
[0069] pl6 Primer F: ACGTCGTGAG...
Embodiment 2
[0126] Embodiment 2, primer and probe design
[0127] In order to optimize the detection reagents suitable for simultaneous detection of human SHOX2 gene, human RASSF1A gene, human ANKRD18B gene and human MPDZ gene methylated DNA, the inventors conducted in-depth research on each gene sequence, after repeated research and comparison, selected The sequence of the amplified region of each gene is shown in Table 2.
[0128] Table 2
[0129]
[0130]
[0131]
[0132]
[0133] According to the regions determined in Table 2, the inventors further optimally designed specific primers and detection probes. The designed primers and detection probes are shown in Table 3. All primers and probes were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
[0134] table 3
[0135]
[0136]
Embodiment 3
[0137] Embodiment 3, different primer probe combination experiments
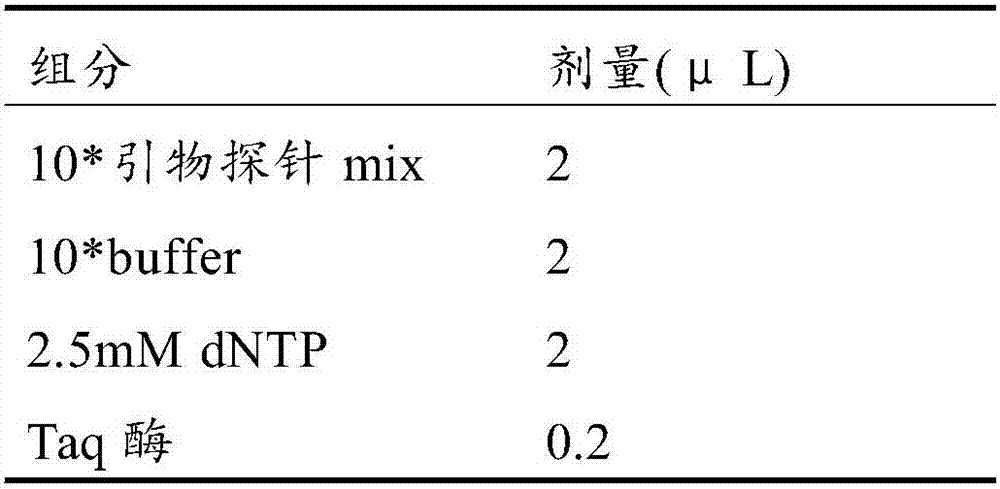
[0138] DNA extraction, sulfite modification, detection system preparation, and PCR amplification procedures were the same as in Example 1, and the amplification objects were modified sample DNA and control DNA (DNA fragments of SHOX2, RASS1FA, ANKRD18B, and MPDZ). After the experiment runs, the analysis steps are as follows:
[0139] (1) Determine whether the experiment is credible:
[0140] (a) There is signal in CY5, and the Ct value of CY5 signal is ≥12, the experiment is credible;
[0141] If the Ct value is less than 12, it indicates that the added DNA is too much, and the DNA should be diluted before doing it;
[0142] (b) If there is no CY5 signal, it indicates that the added DNA contains PCR inhibitors or the DNA treatment fails, and the DNA needs to be re-extracted and treated with sulfite;
[0143] Among them, the positive quality control product reaction tubes should have signals of FAM, VIC an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com