Dual PCR primer, detection method and kit for detecting porcine mouth-foot disease virus and Seneca valley virus
A porcine foot-and-mouth disease virus and detection kit technology, which is applied in the field of animal disease detection, can solve the problems that it is difficult to accurately distinguish between Seneca Valley virus infection and foot-and-mouth disease virus infection, complicated operations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] 1. Preparation of porcine foot-and-mouth disease virus and Senega valley virus nucleic acid templates
[0036] 1) Take clinical samples and freeze and thaw them three times at -80°C for extraction of viral nucleic acids. Total virus RNA was extracted according to the Trizol method.
[0037] 2) Prepare cDNA from the total virus RNA extracted in the above step 1) with a reverse transcription kit, and store it at -80°C for future use. The reaction system is as follows:
[0038] first step
[0039] Random primer 1 μL
[0040] RNA 5.75 μL
[0041] Water bath at 70°C for 10 min, then ice bath for 2 min to obtain the RNA primer mixture
[0042] second step
[0043]
[0044] Water bath at 30°C for 10 minutes, water bath at 42°C for 1 hour, and cool in an ice bath. After reverse transcription, the synthesized cDNA was placed on ice for subsequent experiments or stored at -20°C.
[0045] 2. Screening of specific primers and system optimization of double PCR method for por...
Embodiment 2
[0065] Performance verification of the kit of the present invention
[0066] 1) Specificity test
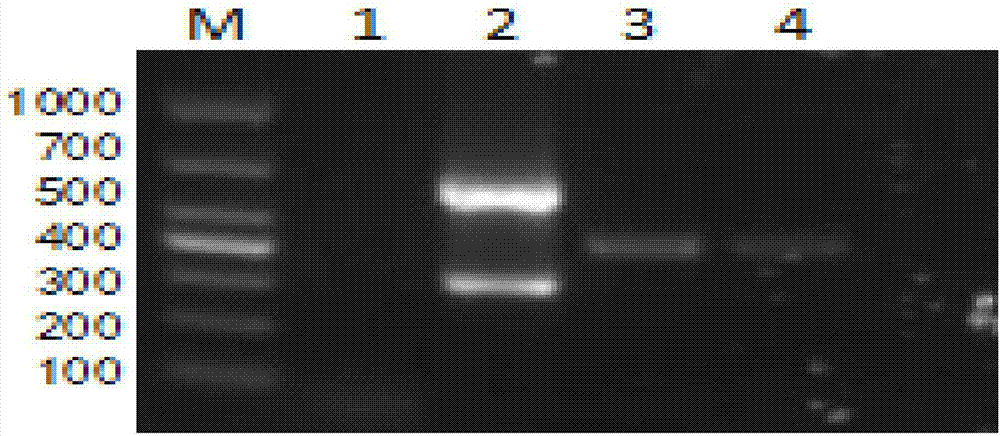
[0067] Porcine foot-and-mouth disease virus (FMDV), porcine senega valley virus (SVV), porcine epidemic diarrhea virus (PEDV), porcine vesicular disease virus (SVDV), vesicular stomatitis virus ( VSV), porcine blue ear disease virus (PRRSV), porcine circovirus (PCV), and swine fever virus (CSFV) were amplified, and sterilized water was selected as a negative control to test the specificity of the established method.
[0068] The result is as Figure 4 As shown, the target fragment size of porcine foot-and-mouth disease virus (FMDV) is 312bp; the target fragment size of porcine senega valley virus (SVV) is 599bp; , respectively 312bp and 599bp; PEDV, SVDV, VSV, PRRSV, PCV, CSFV virus no specific band amplification.
[0069] 2) Sensitivity test
[0070] a) Single-plex RT-PCR sensitivity test
[0071] The plasmid (10 times) diluted with the mixed plasmid of SVV and FMDV respect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com