Mitochondria PCR-RFLP identification method for Crassostrea nippona and Crassostrea gigas
A PCR-RFLP and identification method technology, applied in the field of identified mitochondrial COI gene primers, can solve the problems of difficulty in developing new species of rock oyster culture, difficult to distinguish juveniles, etc., and achieves rapid detection, low equipment requirements, and cost saving. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
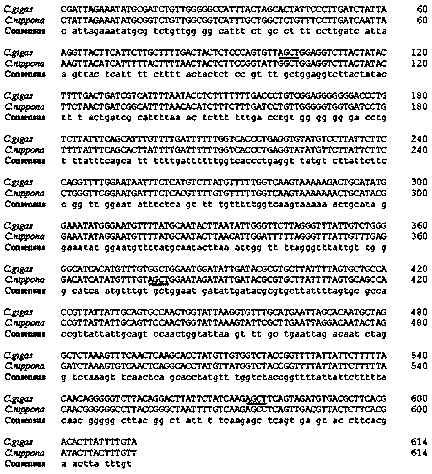
[0020] The present invention is further explained and illustrated by specific embodiments below.
[0021] The rock oyster and long oyster used in this embodiment were taken from the Shandong Rongcheng breeding base. 18 individuals of each type of oyster, a total of 36 individuals.
[0022] Log in to the National Center for Biotechnology Information (NCBI, http: / / www.ncbi.nlm.nih.gov / ) to download the existing rock oyster and rock oyster mitochondrial COI sequence (Rock oyster Genbank accession number: HM015198, rock oyster Genbank accession number is: AB033687).
[0023] Use Mega6.0 software to compare the downloaded sequences.
[0024] Use the primer design software Primer Premier 5 to design primers in the relatively conservative regions of the sequence. The designed COI primer sequence is:
[0025] COIY-F: 5'-CTATTAGAAATATGCGGTCTG-3',
[0026] COIY-R: 5'-AACAAAGTAAGTATCGTGAAG-3'.
[0027] Use phenol-chloroform method to extract DNA from rock oyster and rock oyster, and dilute it to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com