Method For Predicting Organ Transplant Rejection Using Next-Generation Sequencing
A next-generation sequencing and organ technology, applied in the direction of biochemical equipment and methods, instruments, measuring devices, etc., can solve a lot of time and cost problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
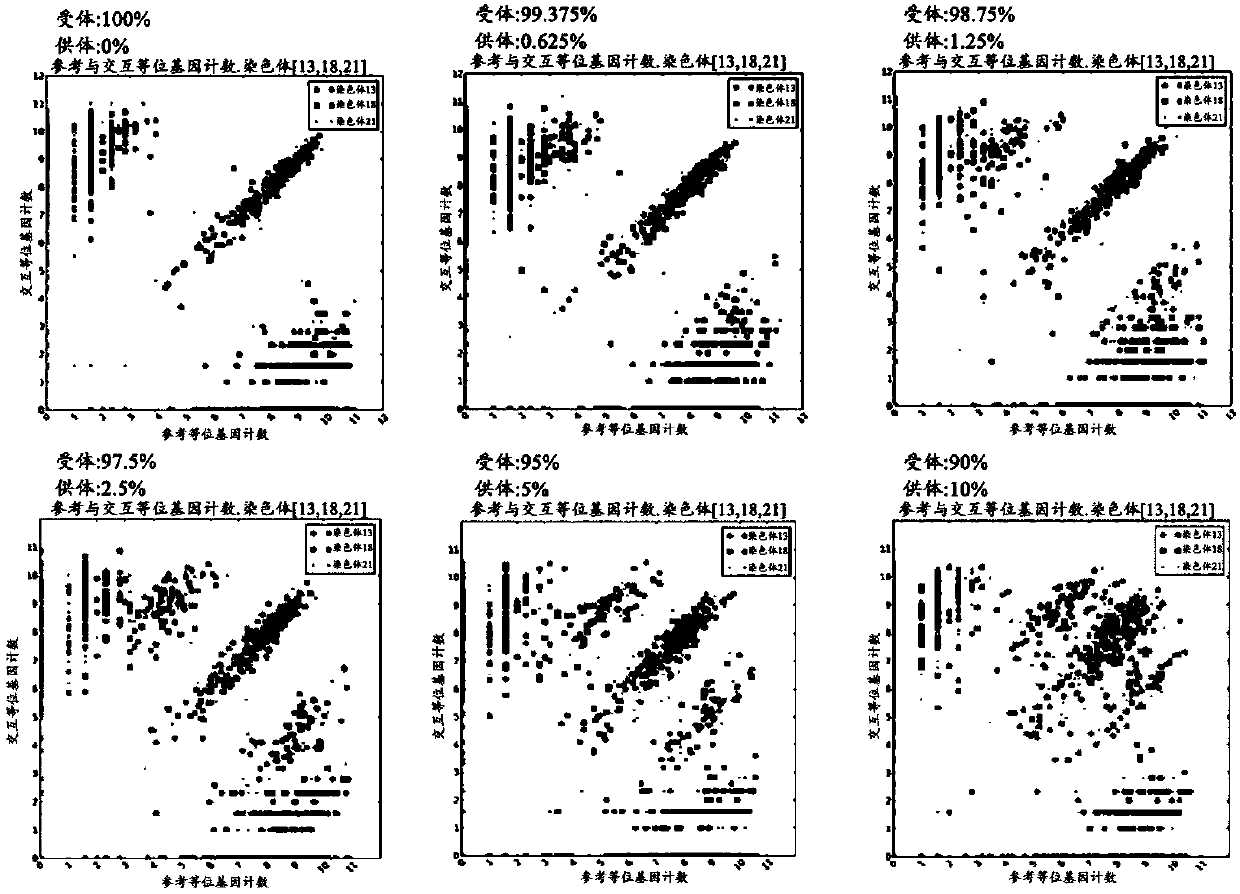
[0125] Example 1: Predicting Organ Transplant Rejection in Artificially Generated Organ Transplant Recipients
[0126] 1.1: Pretreatment for preparation and analysis of artificial DNA samples from organ transplant recipients
[0127] Mixing male DNA (donor) with female DNA (recipient) so that the percentage of male DNA in female DNA is 0%, 0.625%, 1.25%, 2.5%, 5% or 10%, thereby preparing artificial organ transplant patients Genomic DNA samples.
[0128] Custom amplicons were prepared for the TruSeq Custom Amplicon (TSCA) assay (Illumina, USA) using 100 ng of each gDNA. The heat block was adjusted to 95°C, and 5 μl each of DNA and CAT (Custom Amplicon OligoTube) was added to a 1.7-ml test tube. 5 µl each of ACD1 and ACP1 were also prepared as control reagents. Add 40 μl of OHS1 (Oligonucleotide Hybridization Reagent for Sequencing 1) to each tube, mix well using a pipette, maintain each tube at 95°C for 1 minute, then pass it over the oligonucleotide at 40°C Hybridizat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com