High-throughput identification method for genome insulator
A technology of isolators and genomes, applied in the field of genomics, can solve problems that cannot be analyzed on a large scale, cannot be predicted, and cannot be analyzed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
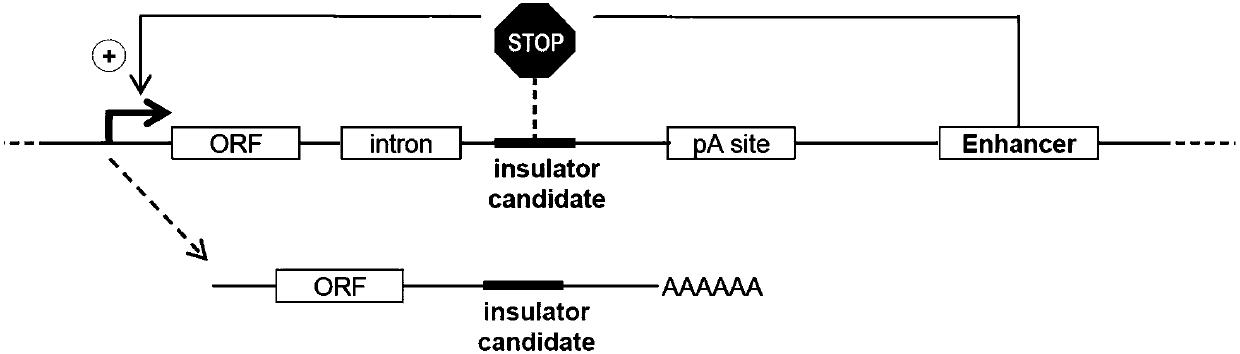
[0029] A method of identifying a genomic isolator comprising:
[0030] 1) Construction of a reporter vector: the reporter vector has a promoter, an ORF region, an intron, an intron candidate sequence insertion region, and an enhancer in sequence; the enhancer is used to enhance the expression of the upstream sequence, and the isolator is inserted into the isolator candidate sequence insertion region candidate sequence;
[0031] 2) express and transcribe the reporter vector;
[0032] 3) Detecting the post-editing transcription of the transcribed sequence to determine whether the isolator candidate sequence is an insulator.
[0033] As a further improvement of the above method, an RNA stabilizing sequence is also inserted between the insertion region of the insulator candidate sequence and the enhancer. Further, the RNA stabilizing sequence is a poly-A sequence. By adding a protective sequence for stabilizing the RNA, the transcribed RNA can be stabilized for a longer period ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com