Application of miRNA
A hsa-mir-4788, hsa-mir-196b-5p technology, applied in the field of molecular biology, can solve problems such as unclear roles
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
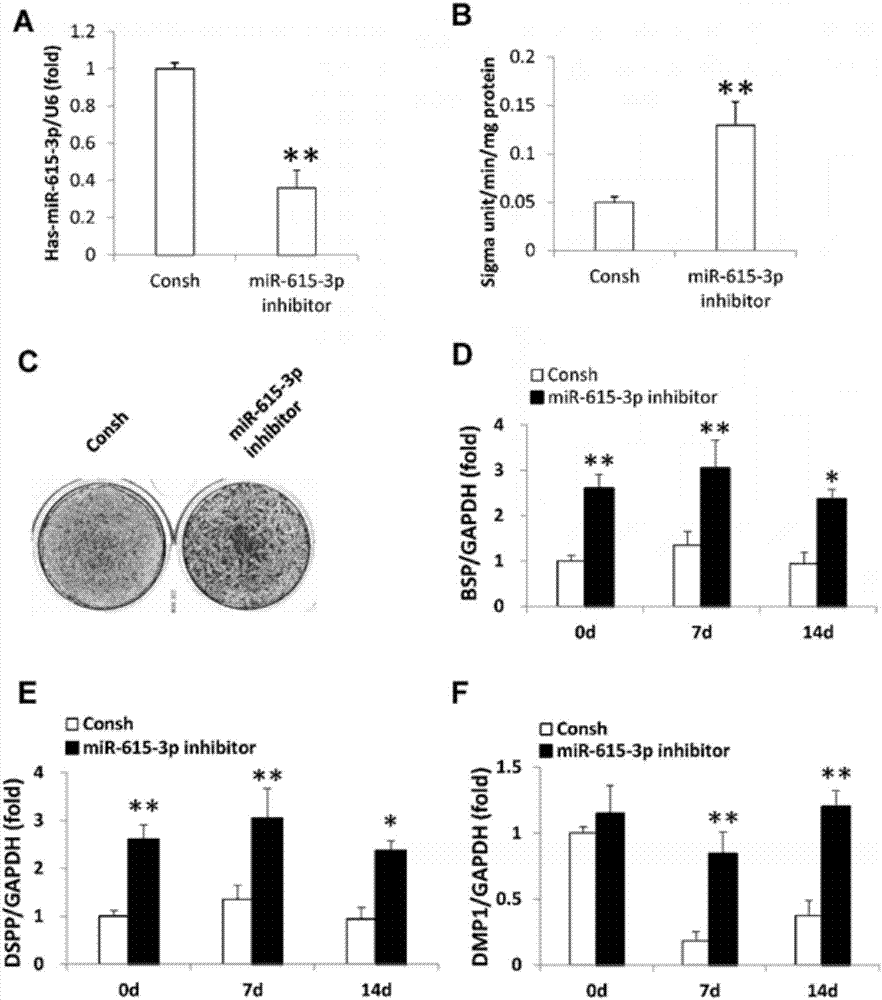
[0045] Example 1 Cell Culture and Osteogenic Differentiation
[0046] All research involving human stem cells adheres to the ISSCR "Guidelines for Conduct in Human Embryonic Stem Cell Research". Dental tissues were obtained with the consent of the patients and strictly followed the guidelines of the Beijing Stomatological Hospital Affiliated to Capital Medical University. Teeth were first sterilized with 75% alcohol and then rinsed with phosphate buffered solution. Periodontal ligament stem cells, dental pulp stem cells and apical papilla stem cells were isolated, cultured and identified as previously described. Briefly, apical papilla stem cells were isolated from the apical tooth papilla, periodontal ligament stem cells were isolated from the periodontal ligament in the middle third of the root, and dental pulp stem cells were isolated from the coronal pulp. Next, these stem cells were placed in a solution of 3 mg / ml collagenase I (Worthington Biochemical Corp., Lakewood, ...
Embodiment 2
[0048] Example 2 total RNA isolation and microarray hybridization
[0049] Three groups of total RNA samples from periodontal ligament stem cells, dental pulp stem cells, apical papilla stem cells, umbilical cord mesenchymal stem cells, bone marrow mesenchymal stem cells and adipose mesenchymal stem cells were obtained by using extraction reagents and RNA extraction. Extraction kits (Qiagen, Germany) were presented separately. Total RNA was quantified by a spectrophotometer ND-2100 (Thermo Fisher), and RNA integrity was assessed using an Agilent 2100 (Agilent Technologies). Each group of total RNA samples were further purified using RNA extraction kit and RNase-Free DNase Set (QIAGEN, GmBH, Germany). Microarray analysis used human MiRNAs 4.0 array (Affymetrix). Sample labeling, microarray hybridization, and washing were performed based on the manufacturer's standards. Briefly, total RNA was polyadenylated and subsequently labeled with biotin. Afterwards, the labeled RNA is...
Embodiment 3
[0050] Embodiment 3 biological information analysis
[0051] Bioinformatics analysis was completed (Compass biotechnology Ltd, Beijing, China). Affymetrix Microarray Command Console software (version 4.0, Affymetrix) was used to analyze microarray images to obtain raw data and then provide normalization for receptor modulation analysis. Then, R software was used to continue the data analysis below. Identification of differentially expressed miRNAs by fold changes. The target genes of differentially expressed miRNAs are the intersection points predicted by five databases (miRanda, miRDB, miRTarbase, TargetScan, and miRecords). KEGG analysis was used to determine the role of these target genes. Hierarchical clustering was used to reveal distinguishable expression patterns of miRNAs across samples.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com