Method for detecting methylation of brain glioma MGMT promoter gene
A technology for glioma and detection methods, applied in biochemical equipment and methods, microbe measurement/inspection, etc., can solve problems such as inconvenient operation and poor detection accuracy, and achieve convenient operation, good sensitivity, and high detection accuracy Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
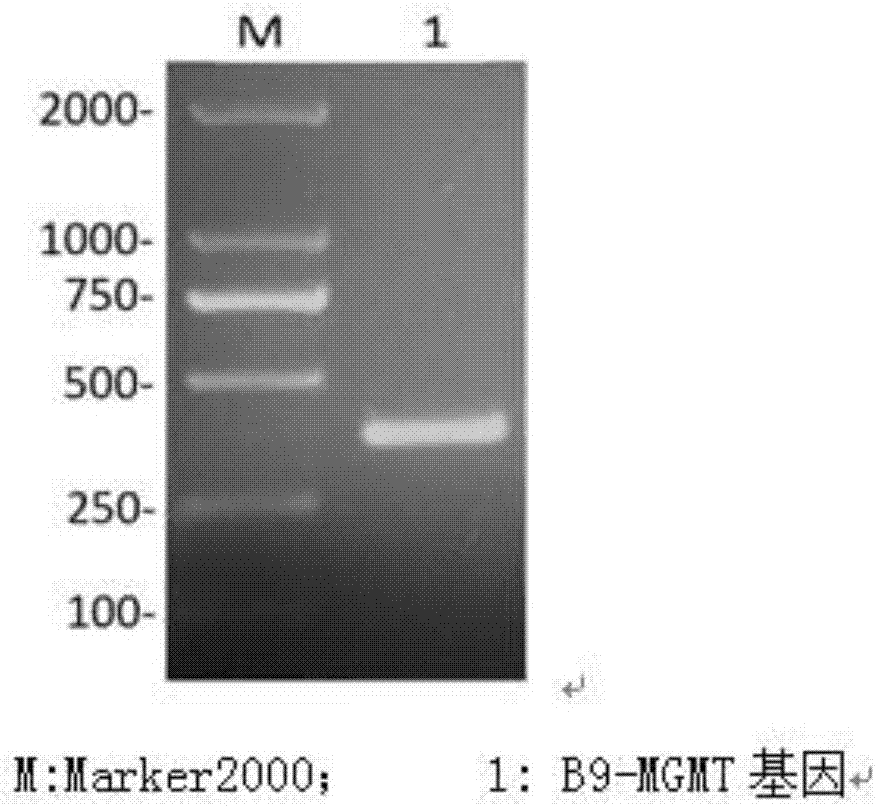
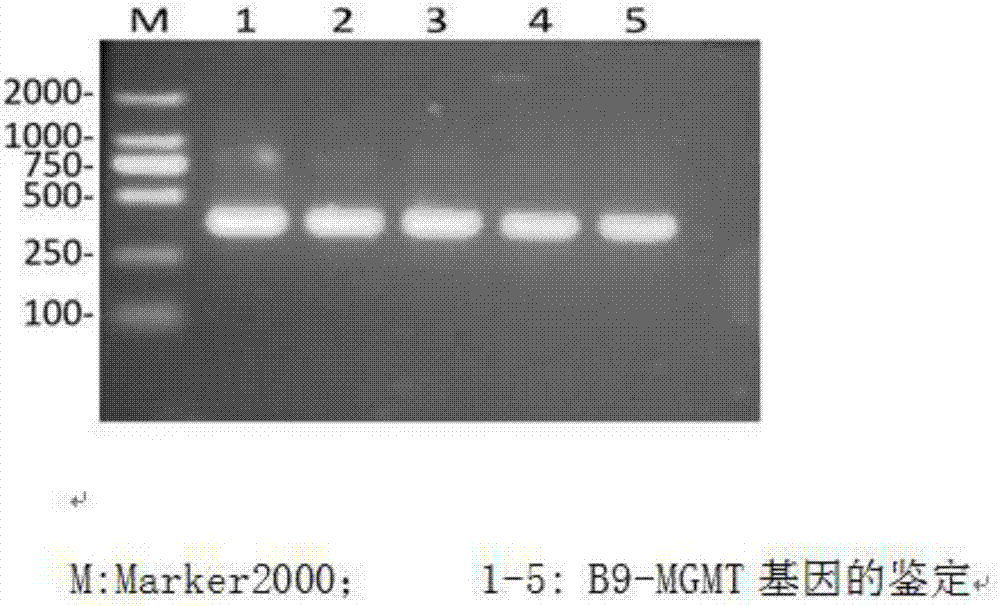
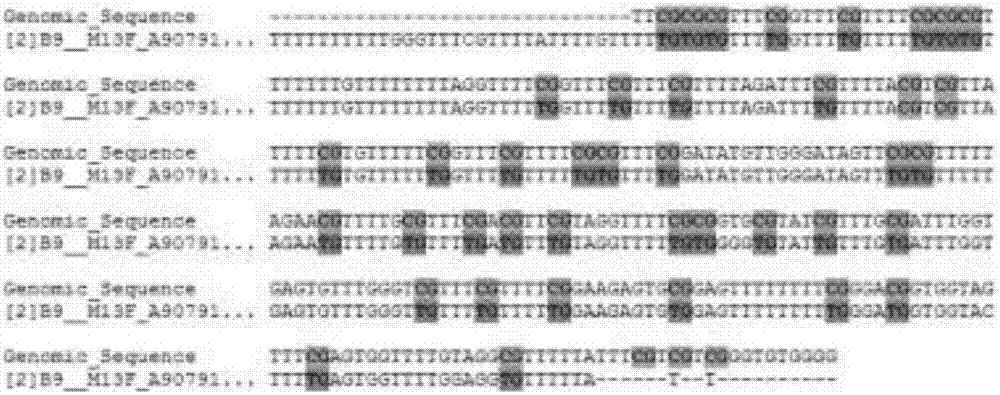
[0043] MGMT promoter methylation detection in patients with glioma A1
[0044] (1) DNA was extracted with a tissue DNA extraction kit from A1 glioma tissue to ensure that the DNA concentration was 100 ng / ul, 260 / 280=1.82.
[0045] (2), take 20ul of DNA and use the methylation modification kit to sulfurize the extracted DNA;
[0046] (3) Use the methylated primers of SEQ NO 1 and SEQ NO 2 to perform drop-down PCR on the sulfur-modified DNA template. The reaction conditions are as follows: pre-denaturation at 95°C for 5 minutes, followed by cycling, denaturation at 95°C for 30s, 57°C Anneal from ℃ to 42℃ for 30s for 2 cycles per degree, finally anneal at 42℃ for 30s for 10 cycles, extend at 72℃ for 45s, a total of 42 cycles, after that, continue to extend at 72℃ for 5min, store at 4℃, the reaction system is 25uL system, It includes: template (sulfurized product): 2uL, SEQ NO 1: 1uL, SEQ NO 2: 1uL, 10×MSP buffer: 2.5uL, 20mM dNTP: 2ul, Hot start taq: 0.5ul, deionized water: 16uL; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com