Methods and compositions for treating C-MET associated cancers
A correlation, cancer technology, applied in the direction of drug combination, medical raw materials derived from bacteria, medical raw materials from fungi, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1: Establishing a FIP- gts Patient-Derived HCC for Preclinical Trials
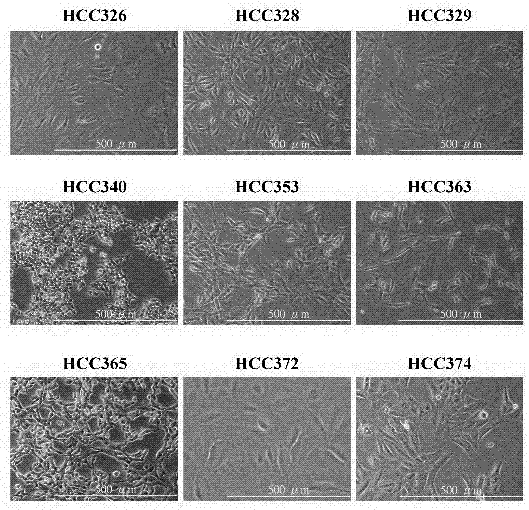
[0073] Clinically derived HCC cell lines were established from a portion of surgically obtained HCC tissue with patient consent and approval by the Buddhist Tzu Chi General Hospital Research Ethics Committee (IRB101-62). Briefly, HCC tissues were pretreated with collagenase, followed by selection of HCC cell lines on mitomycin-treated NIH3T3 feeder layers for a total of 4-6 passages. Obtain homogeneous populations of HCC cells and test their sustained proliferation (over 20 passages) and metastatic potential in vitro and in vivo. Characterization of HCC tumor cell lines was confirmed by detection of HCC tumor markers such as Glypican 3 (GCP3) after more than 40 passages.
[0074] Patient-derived HCC cell lines were established and their phenotypes characterized. Morphologies of nine HCCs (referred to as HCC329, 328, 326 and 340, 353, 365, 363, 372, 274) were shown ( Figure 1A ). Sever...
Embodiment 2
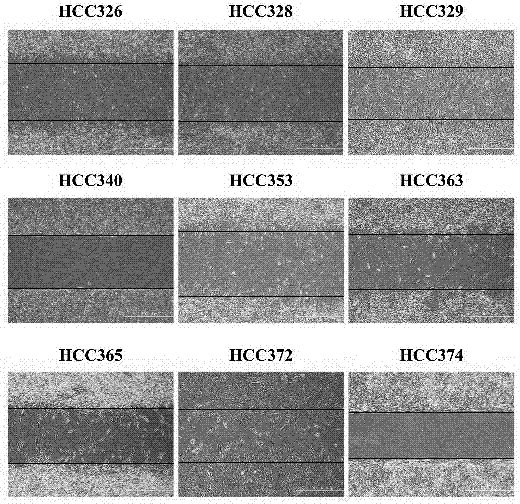
[0075] Embodiment 2: Wound healing migration test
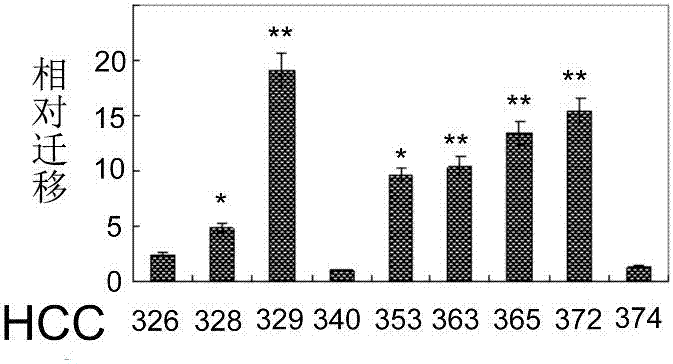
[0076] HCC cells were cultured on 24-well dishes provided with wound healing culture inserts until the cells became confluent, then cells were serum starved for 24 hours, after which the culture inserts were removed. After appropriate processing, pictures were taken using a phase-contrast microscope at the indicated times. The quantitative analysis of the motility of the HCC cell line described in Example 1 was performed by directly counting the cells that migrated to the blank area at 48 hours using the Image J. software taken from the NIH website. result in Figure 1B show. In general, mesenchymal HCCs are more mobile than epithelial HCCs. Among them, mesenchymal phenotypes HCC329 and HCC372 exhibited the highest activity, and epithelial phenotypes HCC340 and HCC374 exhibited the lowest activity ( Figure 1C ).
Embodiment 3
[0077] Example 3: Analysis of signaling in patient-derived HCCs
[0078] The status of key signaling members involved in tumor progression of HCCs, including c-Met, ERK, JNK and AKT, was further examined in the patient-derived cell line established in Example 1 and the commonly used HCC cell line HepG2. Total protein was collected from these cells and subjected to immunoblot analysis using GAPDH as a loading control. Antibodies to p-c-Met, p-JNK, p-ERK, p-paxillin (p-paxillin; S178), GAPDH and ERK were purchased from Santa Cruz Biotechnology (California, USA). Band intensities on the blots were quantified using Image J. software. figure 2 Results shown are representative of 2 reproducible experiments.
[0079] Such as figure 2As shown, c-Met (β subunit, M.W. 140kD) was highly expressed in HCC 372, 340 and HepG2, slightly detected in HCC374, but not observed in other cell lines. Dimerization of c-Met is known to activate the phosphorylation of a tyrosine residue (Tyr1234)...
PUM
| Property | Measurement | Unit |
|---|---|---|
| purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com