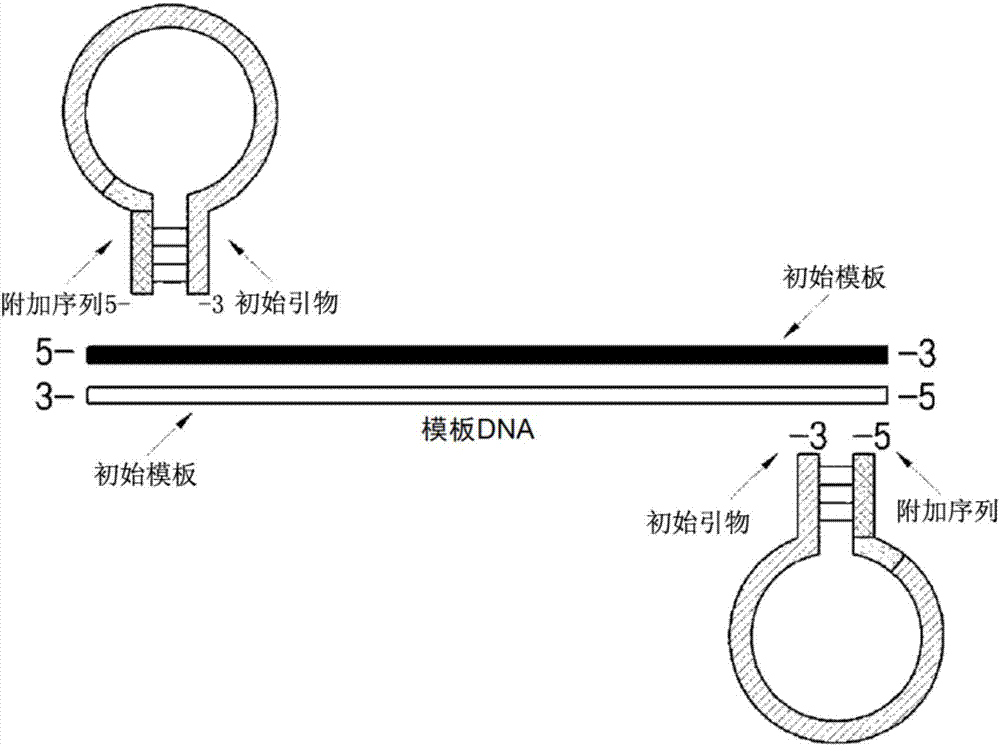
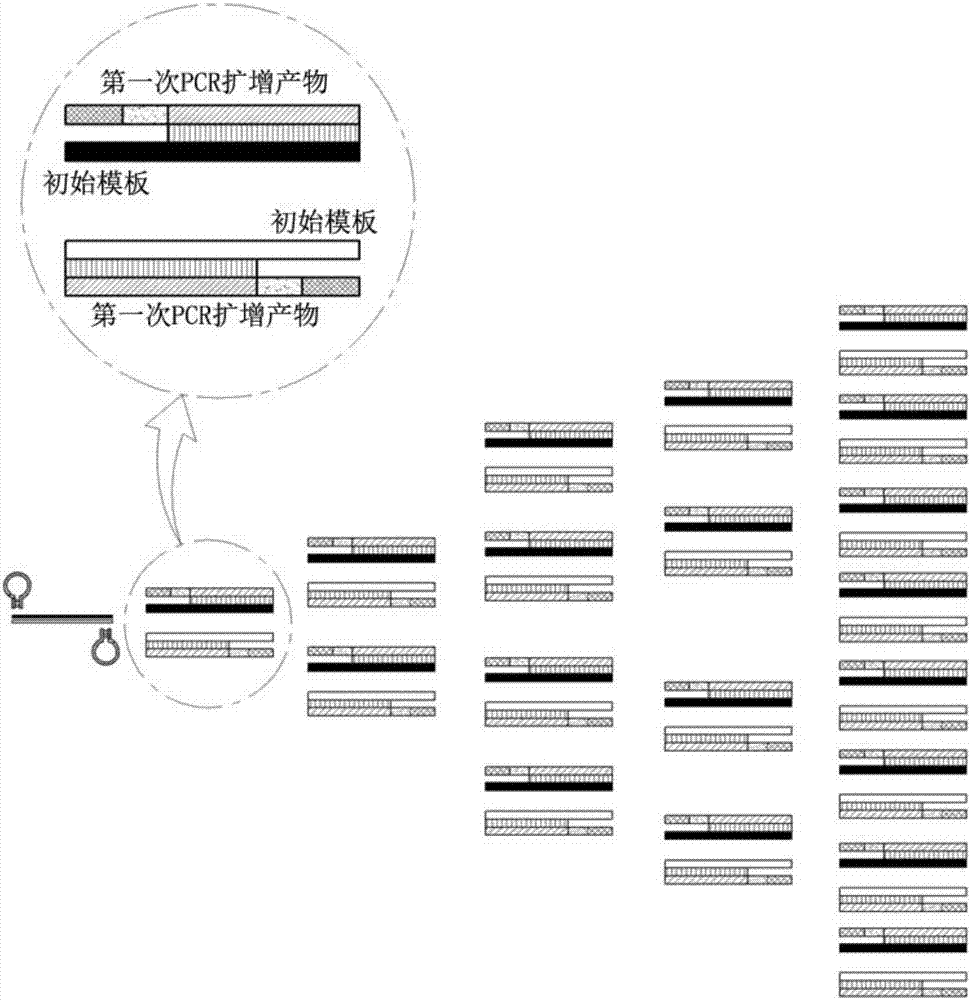
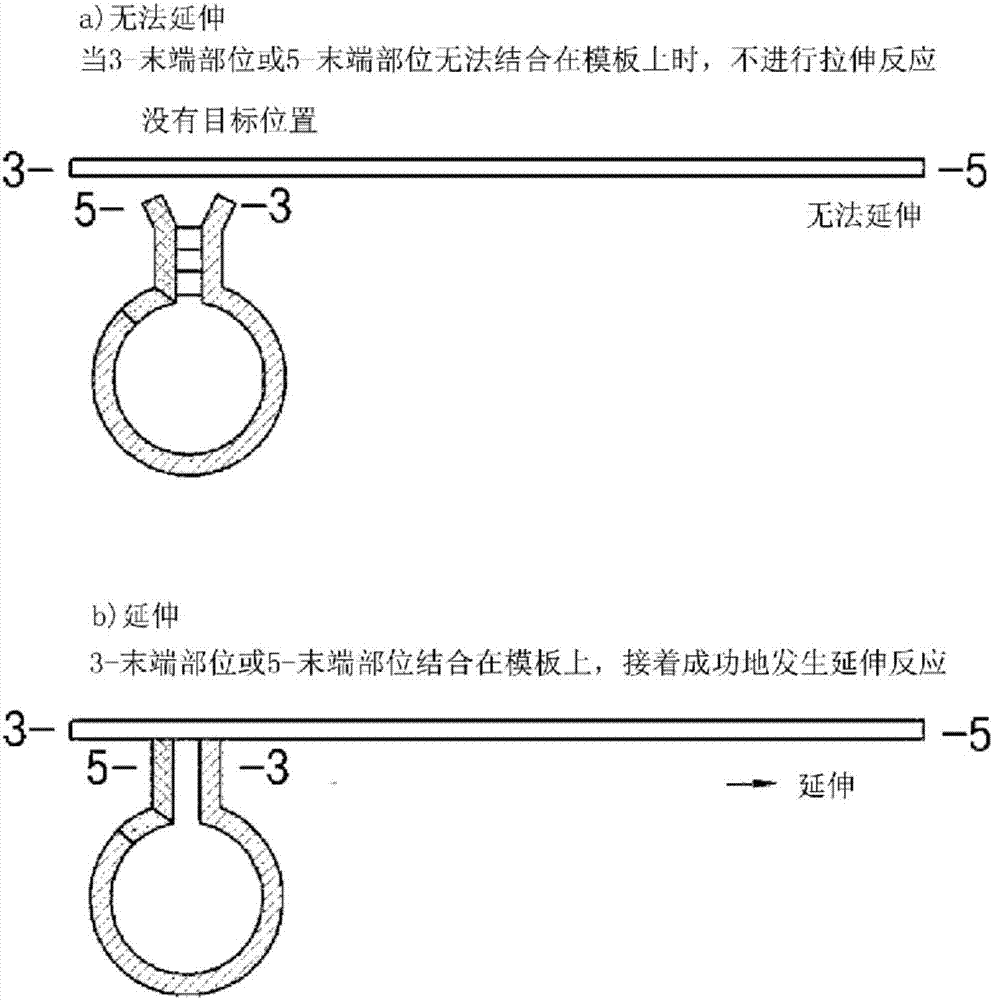
Dumbbell-structure oligonucleotide, nucleic acid amplification primer comprising same, and nucleic acid amplification method using same
A technology of oligonucleotides and dumbbells, applied in the field of dumbbell structure oligonucleotides, can solve the problems of unsure of the application technology and should not complement each other, and achieve easy detection and analysis of single nucleotide polymorphisms, increased sensitivity and effect of specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0121]Example 1: Amplification of Specific Genes for Sexually Transmitted Diseases
[0122] DNA was extracted from samples obtained from 20 patients with suspected sexually transmitted diseases. For the obtained DNA, 2 μl of 10X polymerase chain reaction buffer solution (750 mM Tris-HCl (pH 9.0), 20 mM MgCl 2 , 500mM KCl, 200mM (NH 4 ) 2 SO 4 ), 2 μl of 2.5mM dNTP mixture (2.5mM dATP, 2.5mM dGTP, 2.5mM dTTP, 2.5mM dCTP), 2.0unit of Taq polymerase (Biotools, Spain) and 1 μl of the sequence numbers 1 to 26, 36 and 37 The DS primer (0.5 μM) of the base sequence was added to three distilled water, and after titrating to 20 μl, polymerase chain reaction (10 minutes at 94°C, 30 seconds at 94°C, 60 seconds at 65°C, 60 seconds at 72°C seconds, 35 cycles), the amplification product was obtained. At this time, the base sequences of the respective DSO primers used are shown in Table 1 below. For reference, in the nucleotide sequences recorded in the sequence number 1 to 37 on the se...
Embodiment 2
[0139] Embodiment 2: Utilize the single nucleotide polymorphism analysis of the MTHFR gene of DSO primer, BRAF gene and APC gene
[0140] In order to amplify commercially obtained human MTHFR, BRAF, and APC genes (wild type, hetero type, homo type), human genomic DNA (Invitrogen Inc., USA) was used as a template, and normal After the primers were amplified, the amplified product was confirmed by performing a result confirmation method using restriction enzyme treatment and polymerase chain reaction using the DSO primer of the present invention.
[0141] Table 2
[0142]
[0143] 2μl (50ng / μl) of genomic DNA collected from human blood, 2μl of 10X polymerase chain reaction buffer solution (750mM Tris-HCl (pH 9.0), 20mM MgCl 2 , 500mM KCl, 200mM (NH 4 ) 2 SO 4 ), 2 μl of 2.5mM dNTP mixture (2.5mM dATP, 2.5mM dGTP, 2.5mM dTTP, 2.5mM dCTP), 1.5unit of Taq polymerase (Biotools, Spain) and 1 μl of the mixture of each DSO primer (0.5 μM) Mix, add triple distilled water, titrat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com