Method for analyzing children autism intestinal flora diversity and specific primer pair
A primer pair and specificity technology, applied in the field of biotechnology detection, can solve problems such as deviation of research results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
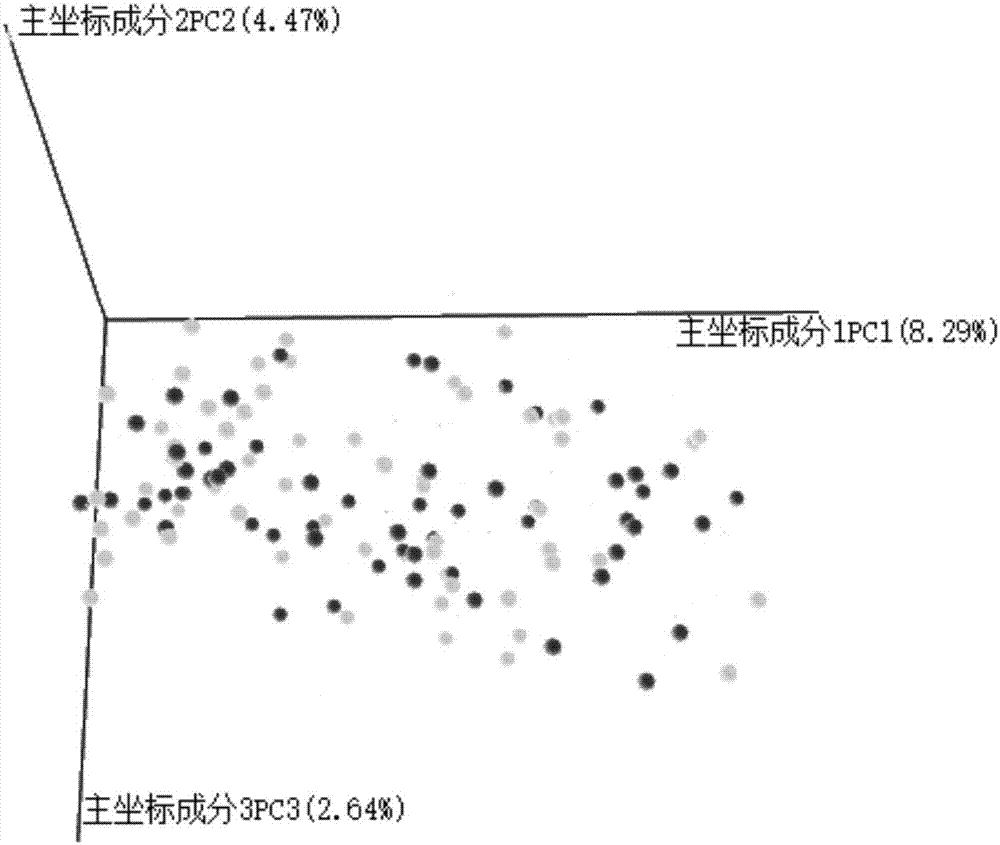
[0089] In Example 1 of the present invention, the samples to be tested are stool samples of autistic children, specifically 30 stool samples of normal children and 59 stool samples of autistic children. A total of 89 disease samples were tested.
[0090] Using the above method, the DNA of the intestinal flora in different patients was extracted, and the extracted DNA was subjected to PCR reaction, and the product of the PCR reaction was subjected to agarose gel electrophoresis, and then the PCR product was purified to construct a library and prepare for Machine sequencing, followed by the following steps:
[0091] 1. Fecal sample sampling method
[0092] Fecal samples were collected in strict accordance with the experimental procedures.
[0093] 2. Extraction method of stool DNA
[0094] Fecal DNA was extracted by CTAB method.
[0095] 3. Determination of DNA content
[0096] The purity of the extracted DNA was detected by a NanoDrop 2000 spectrophotometer, and it was fou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com