Protein-ligand interaction fingerprint spectrum-based drug target prediction method
A fingerprint and target prediction technology, which is applied in the field of drug target prediction based on protein-ligand interaction fingerprints, to achieve the effect of improving the prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
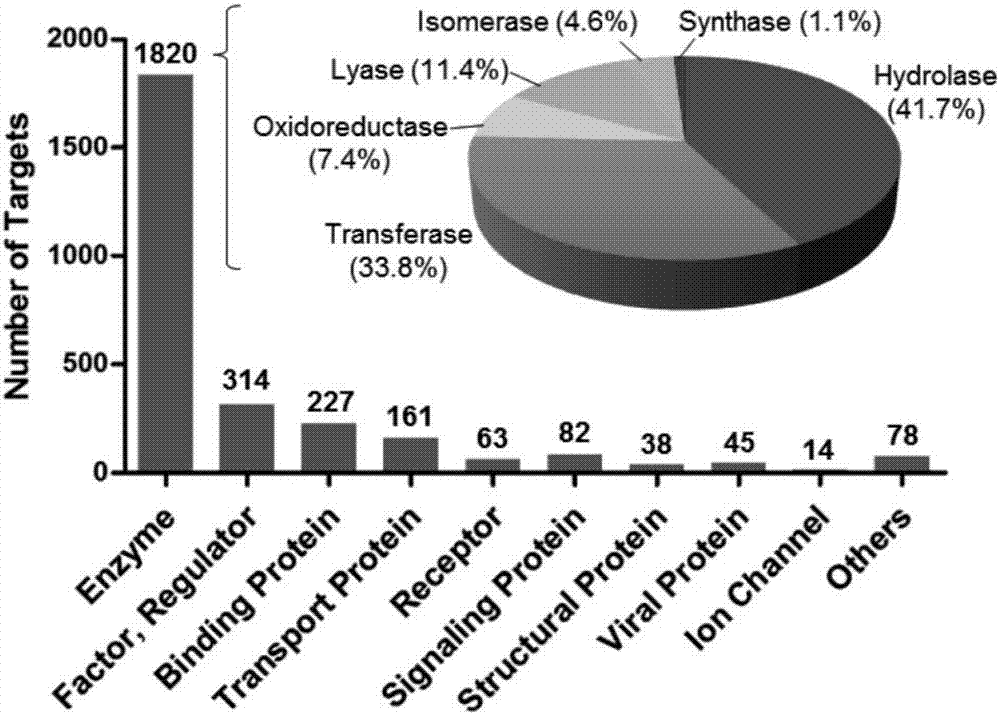
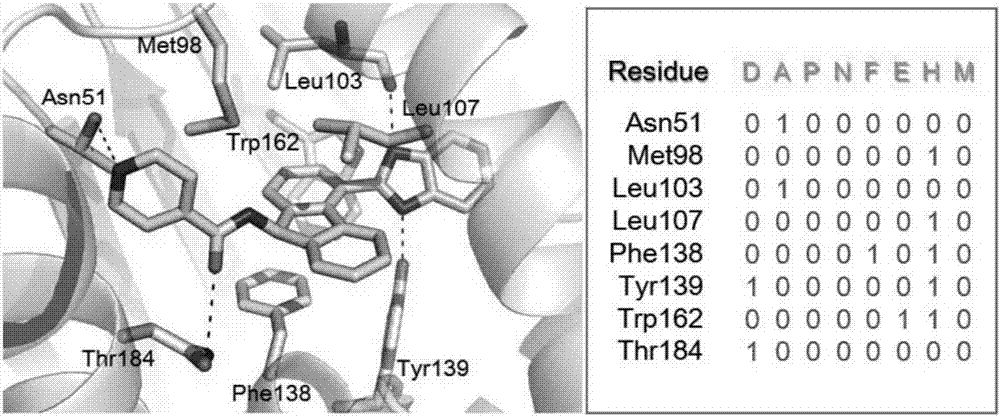
[0039] attached figure 1 A target prediction method based on protein-ligand interaction fingerprints is described. The input medicinal chemical structural formula, which is the optimized three-dimensional structure. According to the target list of the target library, use the constructed program to call the target information sequentially, and call the molecular docking program to compare the input drug three-dimensional structure with the target T i Docking simulation of the active site to generate drug molecules and target T i Molecular docking conformations, in this embodiment there are 10 conformations. Call the built program to perform fingerprint analysis on the molecular docking conformation, and generate the target T i The interaction fingerprint corresponding to each docking conformation of , and calculate the target T i The similarity between the interaction fingerprints of all docking conformations and the reference interaction fingerprints in the fingerprint lib...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com