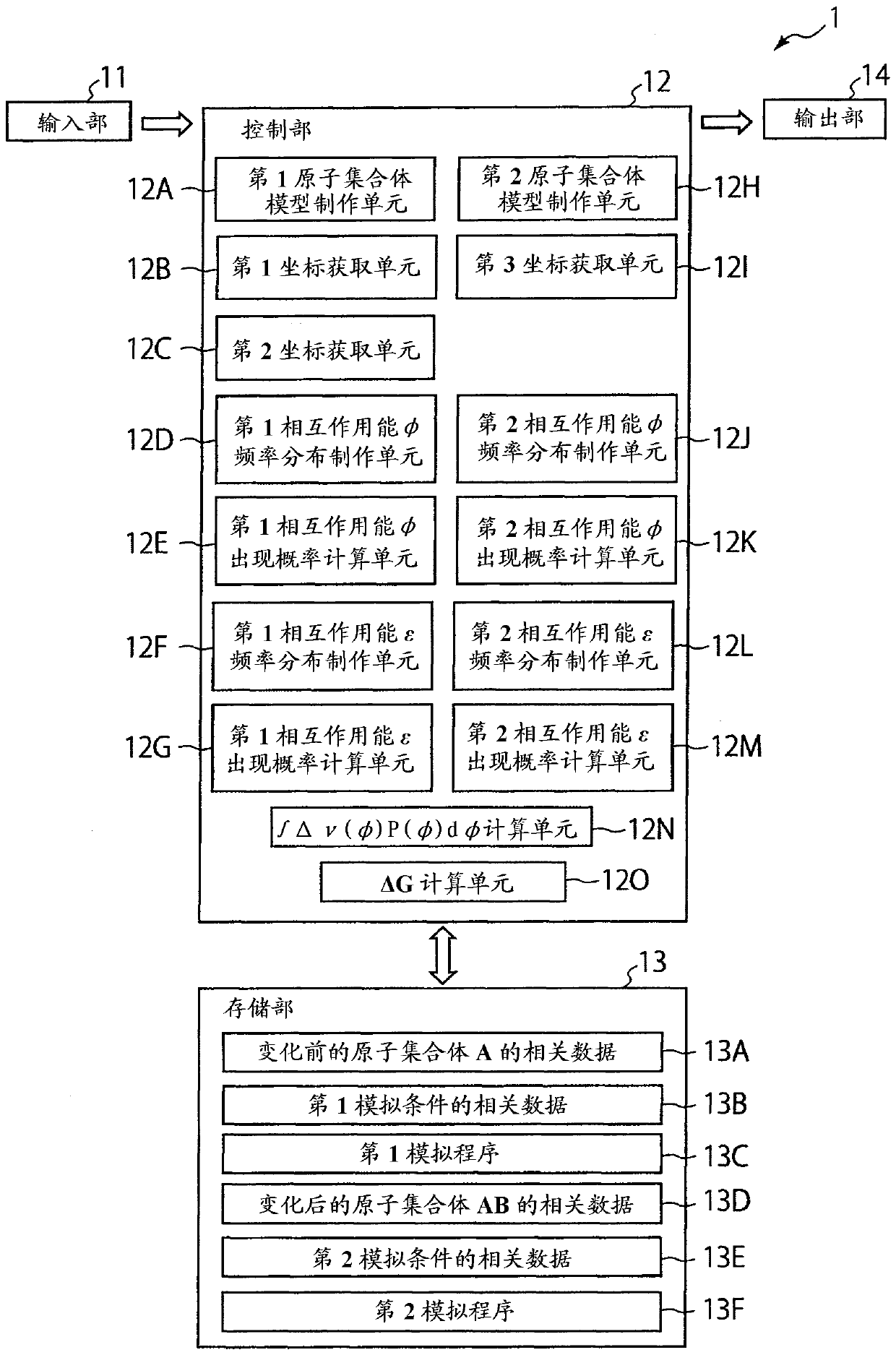
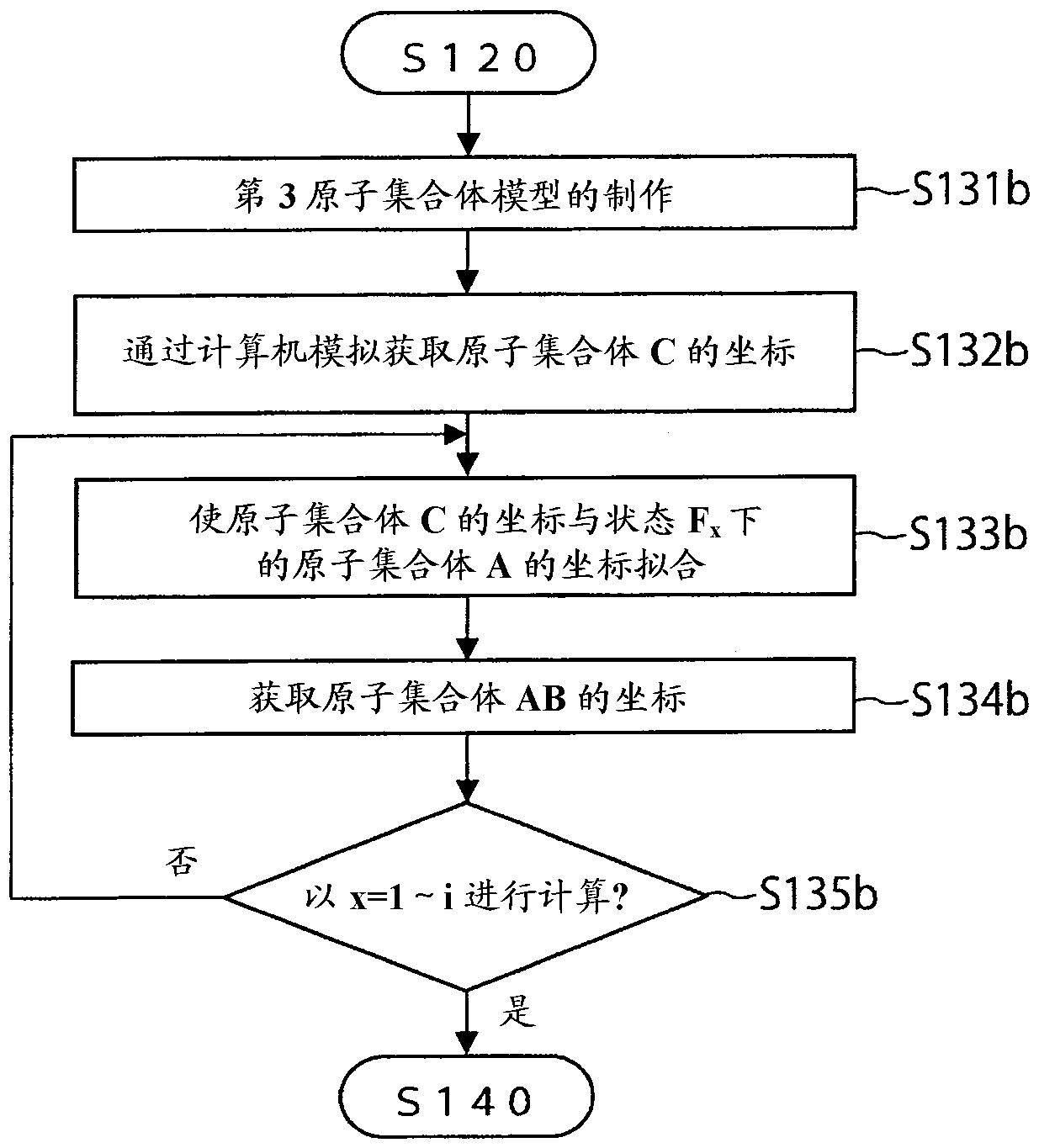
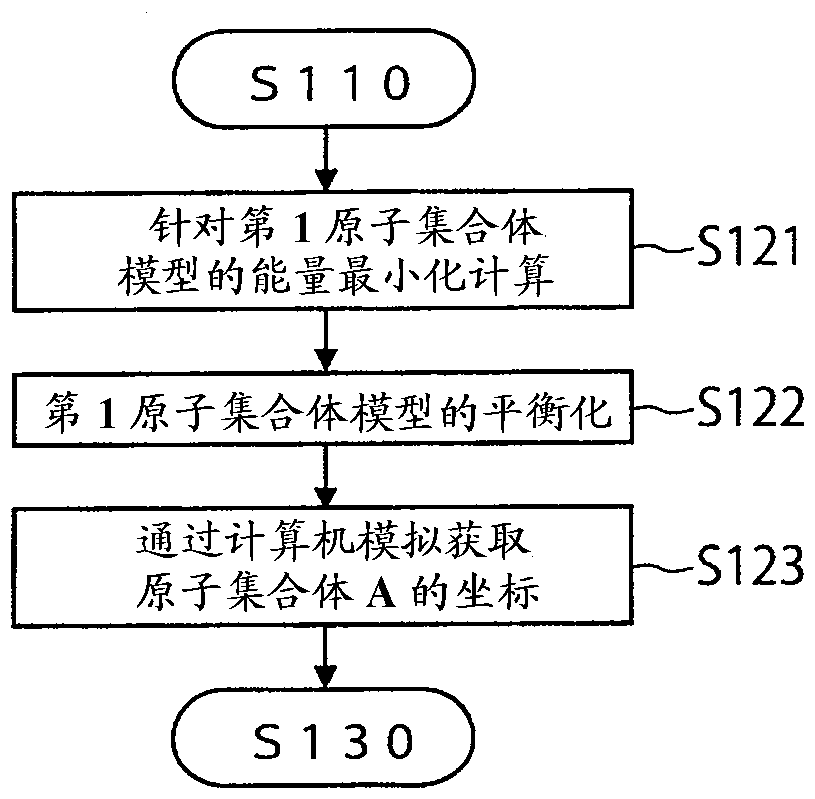
Free energy calculation device, method, program, and recording medium with said program recorded thereon
A computing device and computing method technology, applied in computing, complex mathematical operations, design optimization/simulation, etc., can solve problems such as inability to calculate combined free energy, not a practical computing method, long computing time, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0379] (1) Calculation of ΔG4a
[0380] Regarding the change represented by reaction formula (7), the complex L 0 R and constitute the complex L present in the 0 The atomic assembly before the change of the water molecules around R is called "atom assembly A", and the complex L 1 R and constitute the complex L present in the 1 The changed atomic aggregate of the water molecules around R is regarded as "atomic aggregate AB", and the sum of the free energy of the atomic aggregate A before the change and the free energy of the fragment B, and the value after the change are calculated according to the mathematical formula (1). The free energy difference ΔG4a' of the atomic aggregate AB.
[0381] [chemical formula 5]
[0382]
[0383] It should be noted that since ΔG4a is the amount of free energy change related to the change represented by the reaction formula (8), the value obtained by multiplying the ΔG4a' calculated according to the mathematical formula (1) with a minus ...
Embodiment 2
[0588] Using Ligand L 3 to replace ligand L 2 , performed the same operation as in Example 1, and calculated ΔΔG.
[0589] Ligand L 3 The chemical structure of is as follows.
[0590] [chemical formula 20]
[0591]
[0592] Ligand L 3 The partial charge of each atom of is as follows.
[0593] [chemical formula 21]
[0594]
[0595] The structure of Fragment B is as follows.
[0596] [chemical formula 22]
[0597]
[0598] Here, as described below, ΔΔG was calculated in the same manner as in Example 1 by treating point charges (partial charges; 0.000) as atoms of Fragment B for convenience.
[0599] [chemical formula 23]
[0600]
[0601] The same values as in Example 1 were used for ΔG4a and ΔG3a.
[0602] The calculated ΔG4b was -8.12 (kcal / mol).
[0603] The calculated ΔG3b was -5.45 (kcal / mol).
[0604] The calculated ΔΔG was -0.08 (kcal / mol).
[0605] The ΔΔG recorded in the known literature (Journal of Chemical Theory and Computation, 8, 3686-36...
Embodiment 3
[0607] Using Ligand L 4 Ligand L 2 , performed the same operation as in Example 1, and calculated ΔΔG.
[0608] Ligand L 4 The chemical structure of is as follows.
[0609] [chemical formula 24]
[0610]
[0611] Ligand L 4 The partial charge of each atom of is as follows.
[0612] [chemical formula 25]
[0613]
[0614] The structure of Fragment B is as follows.
[0615] [chemical formula 26]
[0616]
[0617] The same values as in Example 1 were used for ΔG4a and ΔG3a.
[0618] The calculated ΔG4b was -23.00 (kcal / mol).
[0619] The calculated ΔG3b was -19.88 (kcal / mol).
[0620] The calculated ΔΔG was -0.53 (kcal / mol).
[0621] The ΔΔG recorded in the known literature (Journal of Chemical Theory and Computation, 8, 3686-3695 (2012) and Journal of the American Chemical Society, 99, 2331-2336 (1977)) obtained by experiment is -0.40 (kcal / mol), the difference between the ΔΔG calculated by the calculation method of the present invention and the ΔΔG obtai...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com