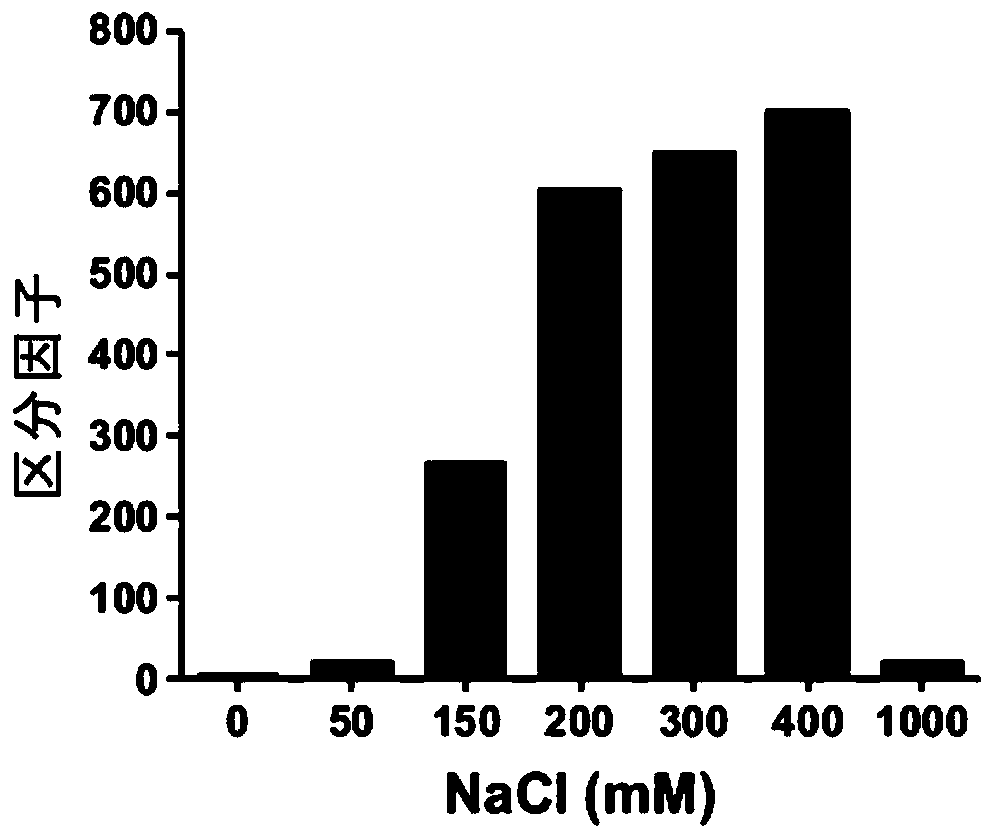
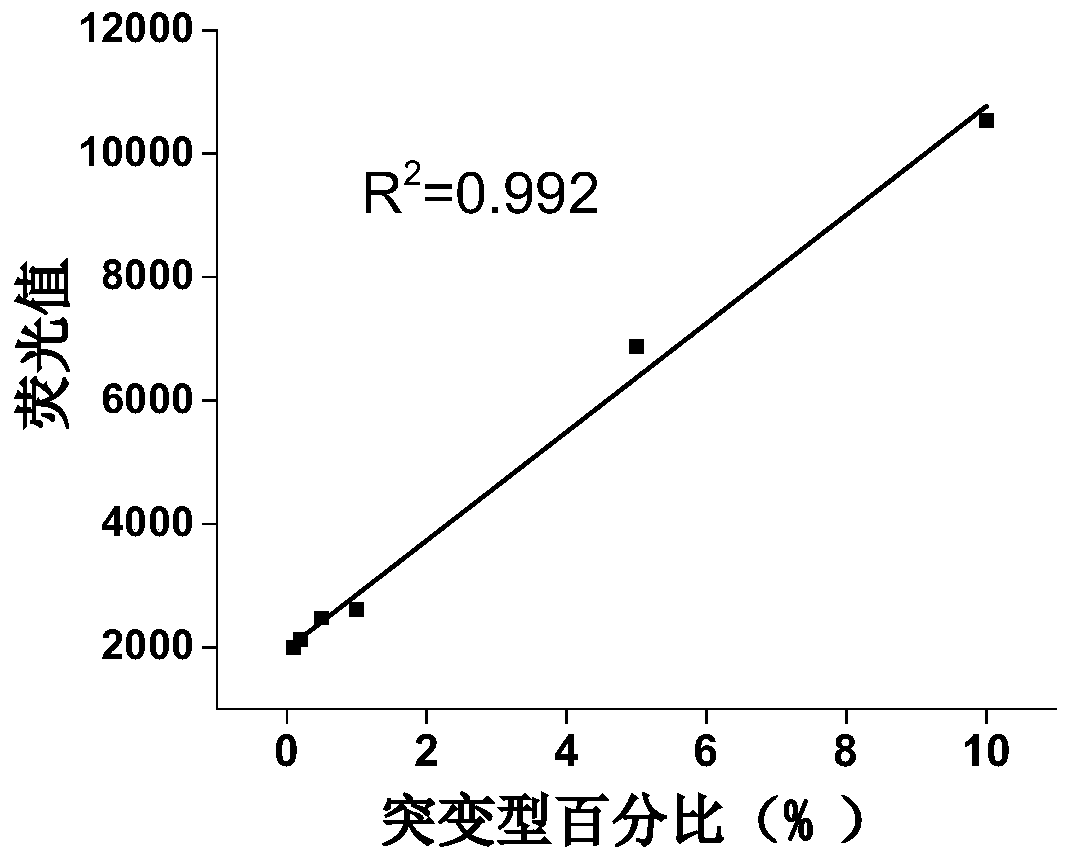
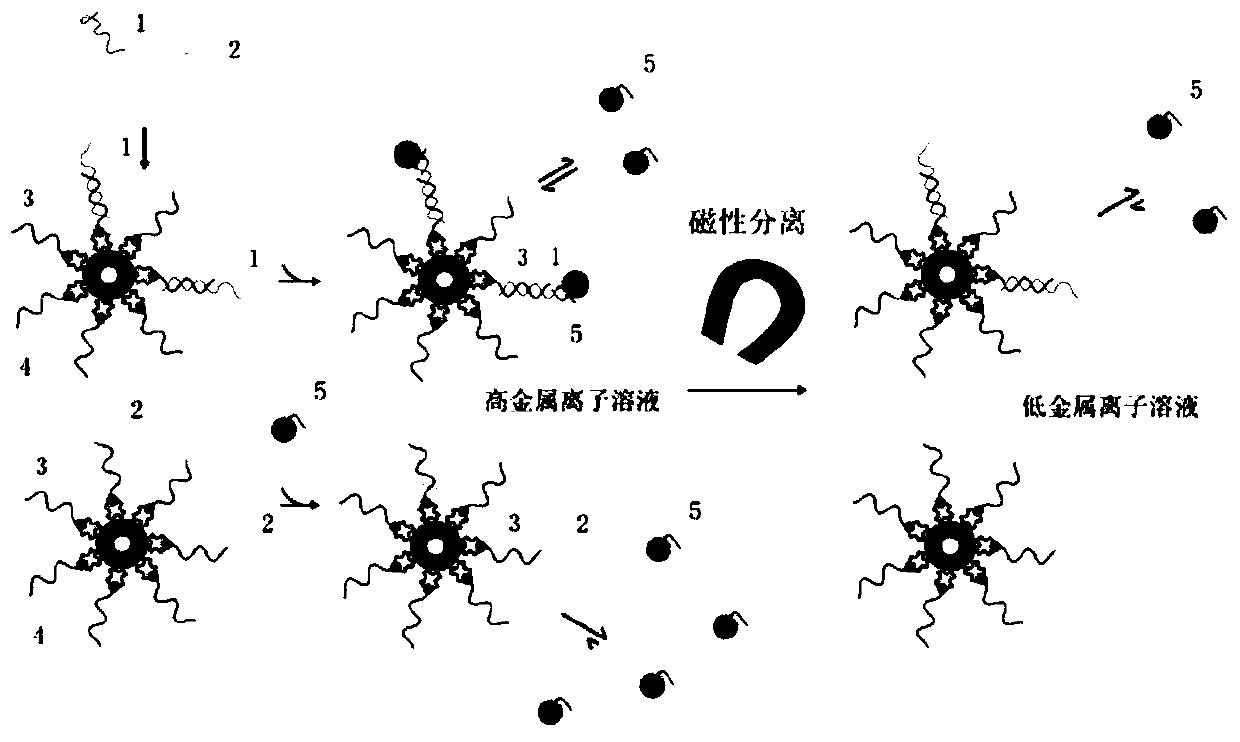
A single base mutation detection method
A single-base mutation and target gene technology, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve problems such as poor performance, and achieve the effects of easy preparation, simple operation, and high repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1: Detection of single base A>G mutation in DNA
[0059] Use a signal probe that is 10 bases complementary to the target gene to detect the A>G gene mutation in the target single-stranded DNA.
[0060] Specific steps:
[0061] (1) Magnetic sphere-coupled biotinylated capture probe
[0062] ①Put the solution of magnetic balls coated with avidin on the surface and vortex for 30s to mix the magnetic balls coated with avidin thoroughly. Pipette 100 μl of magnetic bead solution into a 1.5ml centrifuge tube. Place the centrifuge tube on a magnetic separator, let it stand for 2 minutes, and suck off the supernatant.
[0063] ②Remove the centrifuge tube, add 1ml of buffer I (10mM Tris-HCl (pH=7.5), 1mM EDTA, 1M NaCl, 0.1% Tween-20), vortex and mix well, magnetically separate, and remove the supernatant. Repeat this operation once.
[0064] ③ Add 500 μl of buffer I containing 1.6 μM biotinylated capture probe to make the concentration of the magnetic beads 2 mg / ml, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com