System and method for tumor heterogeneity assessment
A heterogeneity, tumor technology, applied in biochemical equipment and methods, proteomics, microbial measurement/testing, etc., can solve problems such as difficulty in obtaining metastases, inability to represent the complexity of tumors, and inaccurate analysis results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
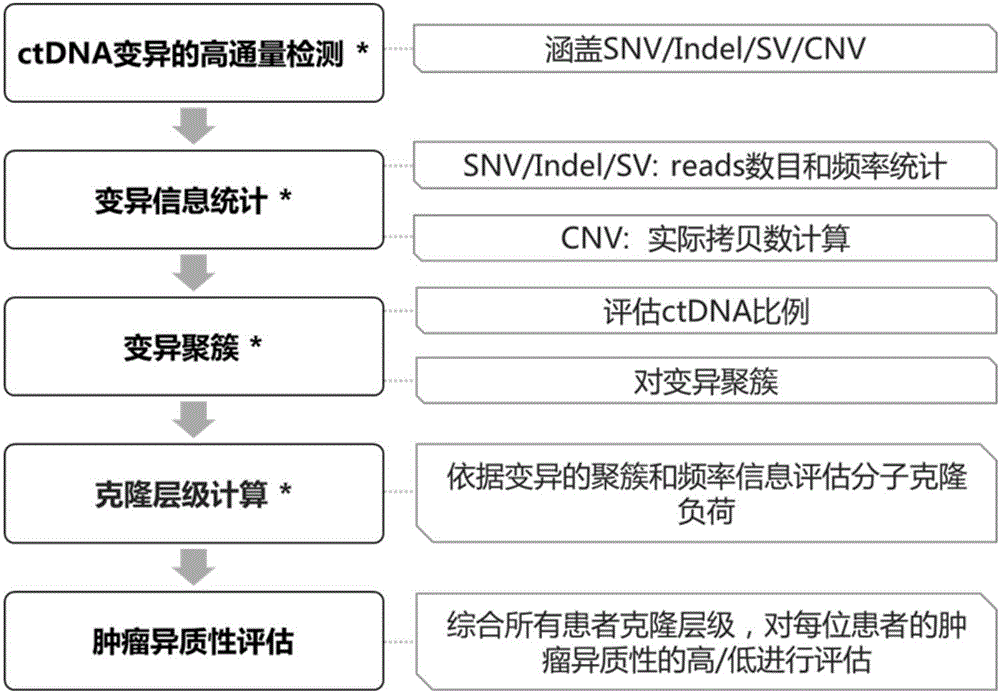
[0131] In this embodiment, 10 cases of lung cancer patients are taken as examples to illustrate the present invention. It should be noted that this embodiment is only for the purpose of illustration, and should not be construed as limiting the application in any way.
[0132] 1. List of variants detected by ctDNA high-throughput sequencing
[0133] 1) Variation V (SNV / indel / SV)
[0134] 2-8 mutations were detected in 10 lung cancer patients, and the detection list of mutation V (SNV / indel / SV) is shown in Table 1.
[0135] Table 1 Variation V (SNV / indel / SV) detection list
[0136]
[0137]
[0138] 2) CNV
[0139] Among the 10 lung cancer patients, only S5 was detected with EGFR amplification, and the amplification factor was 1.73, as shown in Table 2. Therefore, the actual total copy number corresponding to the EGFR Deletion variant detected in S5 was estimated to be 4.
[0140] Table 2 CNV detection list
[0141] Sample serial number Gene copy number v...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com