Fusion protein for screening weak MdmX inhibitor or testing inhibition activity of weak MdmX inhibitor
A technology of fusion protein and inhibitory activity, which is applied in the field of biotechnology and can solve the problems of fluorescein losing its fluorescent function, reducing the accuracy of detection methods, and incompletely consistent molar ratios.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Embodiment 1: p53p-XX-N-MdmX fusion protein expression plasmid construction
[0072] Based on the published crystal structure of the MdmX binding domain p53p of p53 and the p53 binding domain at the amino terminal of MdmX (PDB ID: 3DAB) (refer to Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain, Popowicz, G.M., Czarna , A., Holak, T.A. (2008) CellCycle 7:2441-2443), we established the following p53p-XX-N-MdmX fusion protein model.
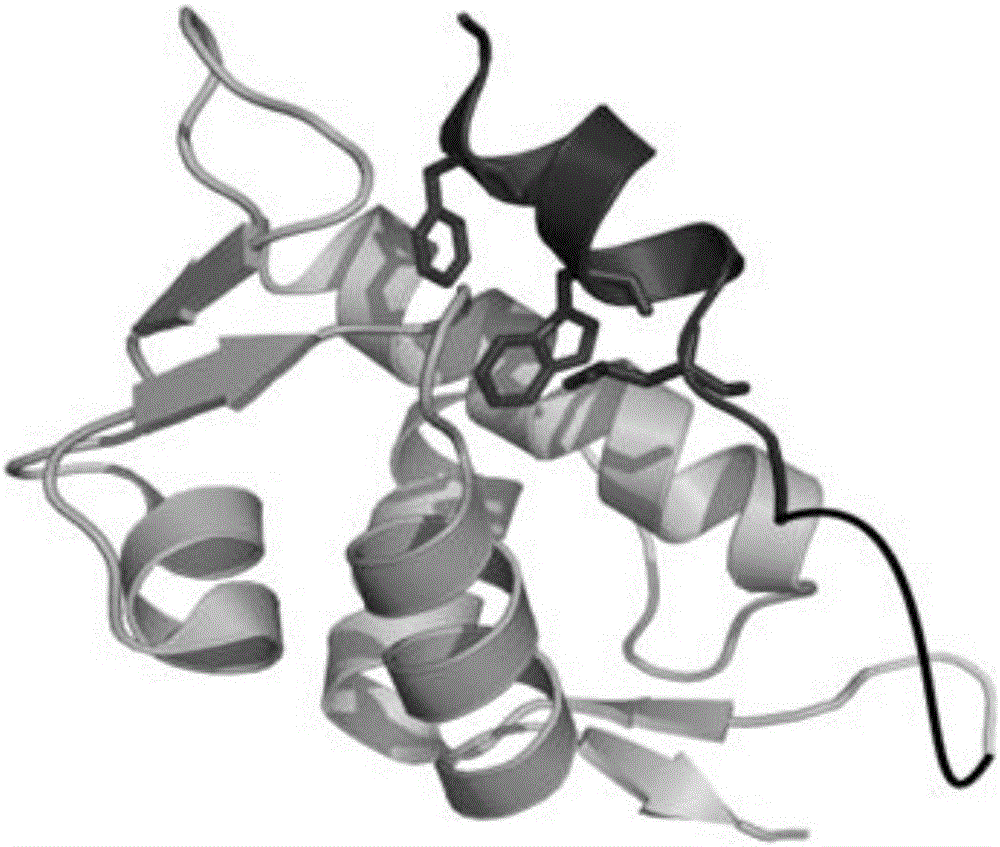
[0073] The amino acid sequence of the p53p-XX-N-MdmX fusion protein is shown below (the tertiary structure of the protein after removing the tag GSSHHHHHHGS is simulated by PymolWin as shown in figure 1 shown):
[0074] GSSHHHHHHGSSQETFSDLWKLLPENGSGSSENLYFQGSQINQVRPKLPLLKILHAAGAQGEMFTVKEVMHYLGQYIMVKQLYDQQEQHMVYCGGDLLGELLGRQSFSVKDPSPLYDMLRKNLVTLAT
[0075] Among them, SQETFSDLWKLLPEN is from the MdmX binding domain sequence of p53, GSQINQVRPKLPLLKILHAAGAQGEMFTVKEVMHYLGQYIMVKQLYDQQEQHMVYCGGDLLGELLG...
Embodiment 2
[0108] Example 2: Expression and purification of p53p-XX-N-MdmX fusion protein
[0109] Transform the pET28a-p53p-XX-N-MdmX plasmid into Escherichia coli BL21(DE3); pick a single colony into 2ml LB K + (Kanamycin) culture medium, 37°C, 200rpm for overnight culture; take 500μl bacterial solution and transfer it to 50ml LB K + Culture medium, 37°C, 200rpm for 3h; transfer 50ml of bacterial solution into 1L LB K + Culture medium, 37°C, 200rpm, until the cell concentration reaches OD 280nm =0.8 or so, add IPTG (final concentration 0.4mM) to induce, and collect the bacteria for about 20 hours (the centrifuge parameters are set to 3500rpm, 30min, 4°C when collecting the bacteria).
[0110] Ultrasonic cell disruptor disrupts cells. According to the ratio of cell volume:buffer volume=1:5, add bufferA buffer solution (50mM Na 2 HPO 4 , 200mM NaCl, 10mM imidazole, 1mM BME (β-mercaptoethanol), pH8.0), the parameters are set as: total time 2min; ultrasonic time 2s; interval time 4s; ...
Embodiment 3
[0112] Embodiment 3: Fluorescence spectrum analysis of p53p-XX-N-MdmX fusion protein model
[0113] Take a frozen 200μl p53p-XX-N-MdmX protein, thaw it quickly, and dilute the protein concentration to OD with phosphate buffer 280nm = 0.1. Take 400 μl to a quartz cuvette, and perform fluorescence scanning on a F-7000 fluorescence spectrophotometer. According to the endogenous fluorescence characteristics of tryptophan, take λ EX 278nm, scan λ EM In the fluorescence intensity in the range of 290nm~500nm, it was found that the maximum fluorescence intensity was at 321nm ( Figure 4 a). Therefore, λ EM 321nm, scan λ EX It is the fluorescence excitation spectrum curve of 245nm~300nm, the result is as follows Figure 4 As shown in b, it can also be known that at λ EX At 278nm, the fluorescence intensity is the largest, so the wavelength of excitation light is set at 278nm.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com