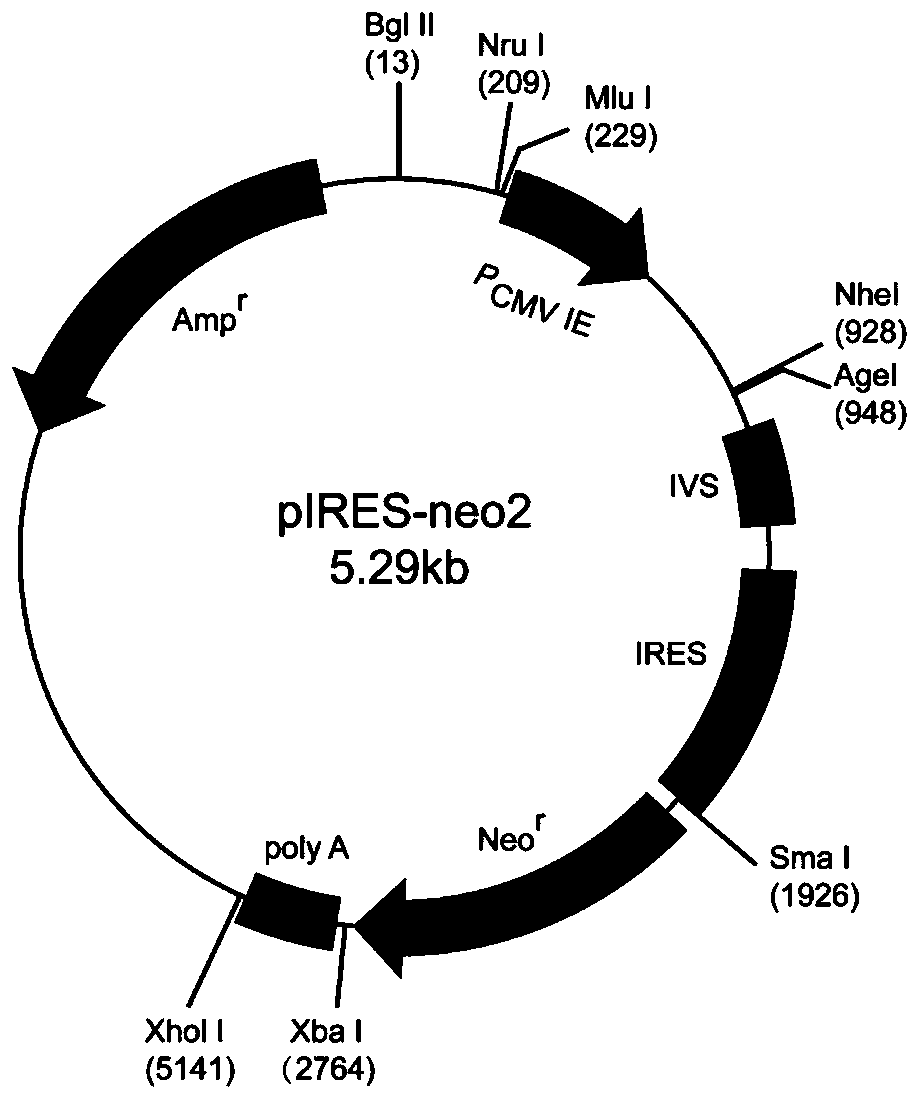
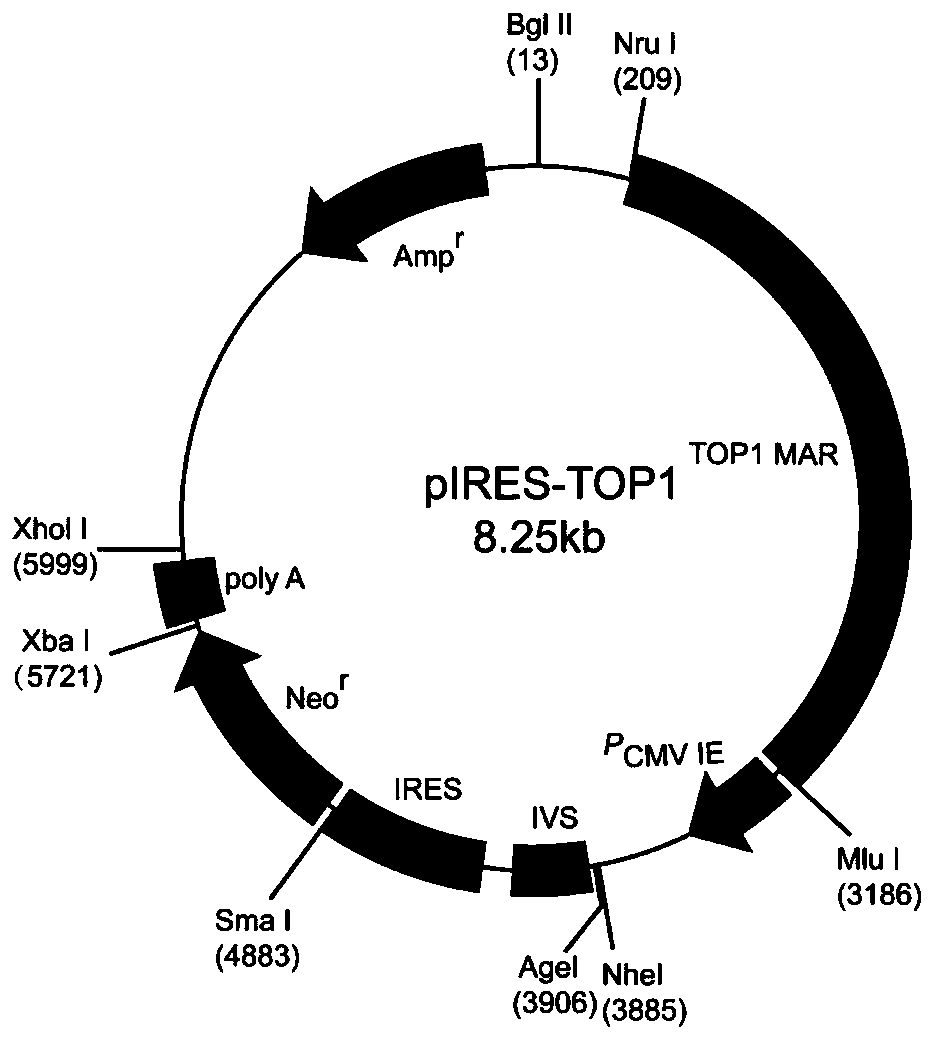
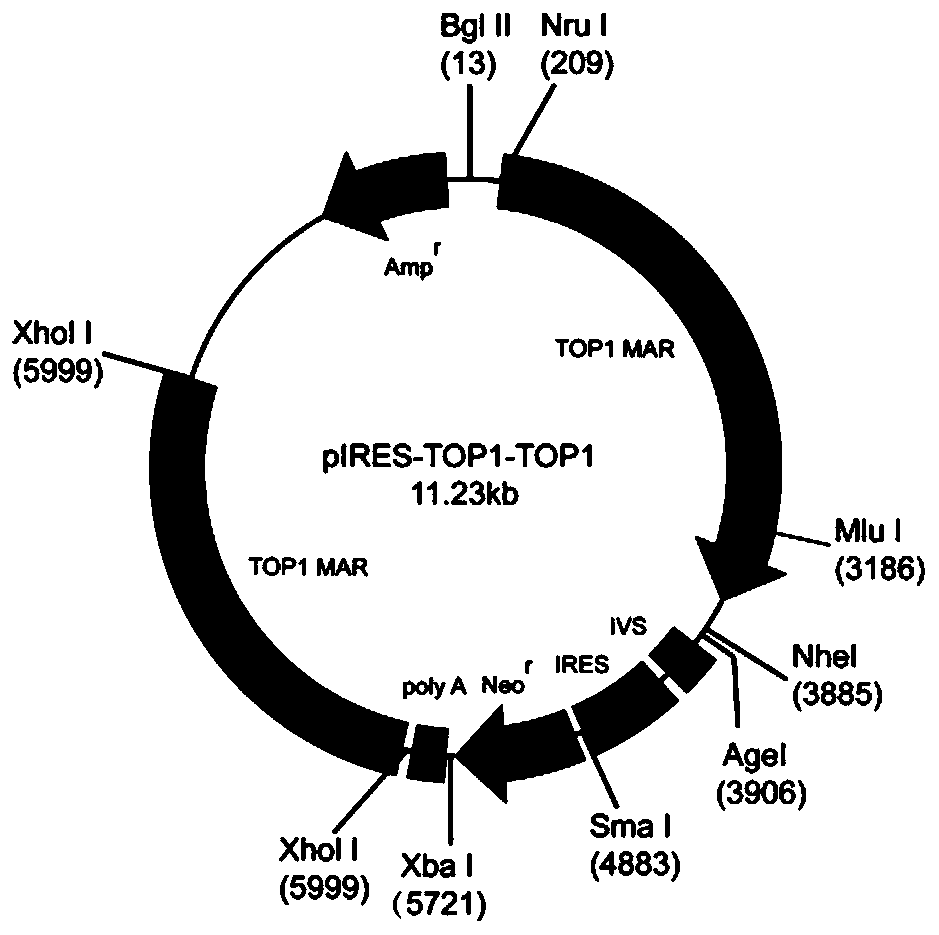
Mammalian cell expression vector, expression system, preparation method and application
An expression vector and mammalian technology, applied in the field of genetic engineering, can solve problems such as the need to improve the expression level of transgenes and long MAR sequences, and achieve the effects of overcoming silencing of transgenes, improving expression levels, and lasting expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] The construction of mammalian cell expression vector pIRES-TOP1, the steps are as follows:
[0041] 1) PCR amplification of TOP1 MAR sequence
[0042] Primers P1 and P2 were designed according to the TOP1 MAR sequence (GenBank: L23999.1, bases 1-2974), and the 5′ ends of the primers were respectively introduced with NruI and MluI restriction sites. The primer sequences are as follows (the underline is the enzyme cutting point):
[0043] P1: 5′-G TCGCGA GGATCCCAATAGGAGTCATT-3′;
[0044] P2: 5′-CG ACGCGT GAATTCACTTCAGGTAACAT-3'.
[0045] Genomic DNA from human peripheral blood was extracted and used as a template for PCR amplification. Primers P1 and P2 were used to amplify the TOP1 MAR sequence. The reaction system is shown in Table 1 below.
[0046] Table 1 PCR amplification system
[0047]
[0048] Reaction program: 95°C for 3min, 94°C for 40s, 56-60°C for 30s, 72°C for 40s, 4 cycles for each annealing temperature, and finally 30 cycles at 55°C for 1min, 72°C...
Embodiment 2
[0056] The construction of mammalian cell expression vector pIRES-TOP1-TOP1, the steps are as follows:
[0057] 1) PCR amplification of TOP1 MAR sequence
[0058] Primers P3 and P4 were designed according to the TOP1 MAR sequence (GenBank: L23999.1, bases 1 to 2974), and the 5′ ends of the primers were respectively introduced with XhoI restriction sites. The primer sequences are as follows (the restriction sites are underlined) point):
[0059] P3: 5′-C CTCGAG GGATCCCAATAGGAGTCATT-3′;
[0060] P4: 5′-C CTCGAG GAATTCACTTCAGGTAACAT-3'.
[0061] Genomic DNA from human peripheral blood was extracted and used as a template for PCR amplification. Primers P3 and P4 were used to amplify the TOP1 MAR gene. The reaction system and reaction conditions were basically the same as in Example 1, and P1 and P2 in Table 1 were replaced with P3 and P4.
[0062] The PCR amplification product was recovered by agarose gel electrophoresis, purified and sent to the biological company for sequ...
Embodiment 3
[0069] The construction of mammalian cell expression system, the steps are as follows:
[0070] 1) PCR amplification of EGFP gene
[0071] Referring to the Enhanced green fluorescent protein (EGFP) gene sequence of the pEGFP-C1 vector (GenBank: U55763.1, bases 613-1332), primers P5 and P6 (used to amplify EGFP gene DNA of 720bp) were designed. The NheI and AgeI restriction sites were introduced into the ' ends respectively, and the primer sequences were as follows (the restriction sites are underlined):
[0072] P5: 5′-CCG GCTAGC ATGGTGAGCAAGGGCGAGGAG-3′;
[0073] P6: 5′-CTA ACCGGT GGACTTGTACAGCTCGTCCATGC-3'.
[0074] Using the pEGFP-C1 plasmid (purchased from Clontech, USA) as a template, primers P5 and P6 were used to amplify the EGFP gene. The reaction system and reaction conditions were basically the same as in Example 1, and P1 and P2 in Table 1 could be replaced with P5 and P6.
[0075] The PCR amplification product was recovered by agarose gel electrophoresis, pu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com