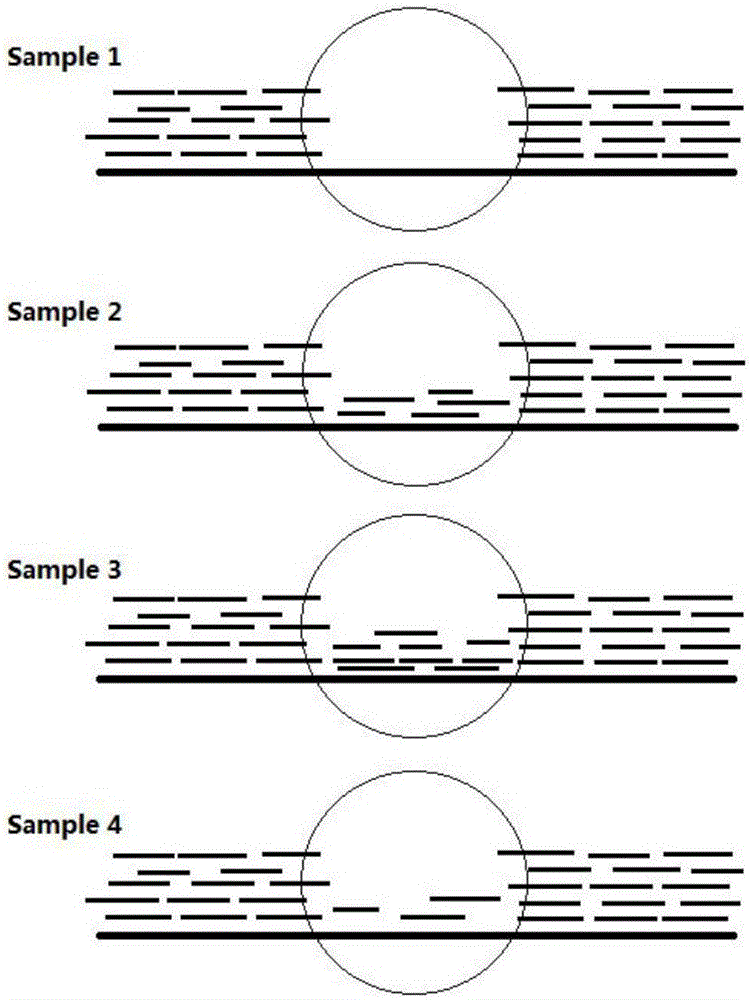
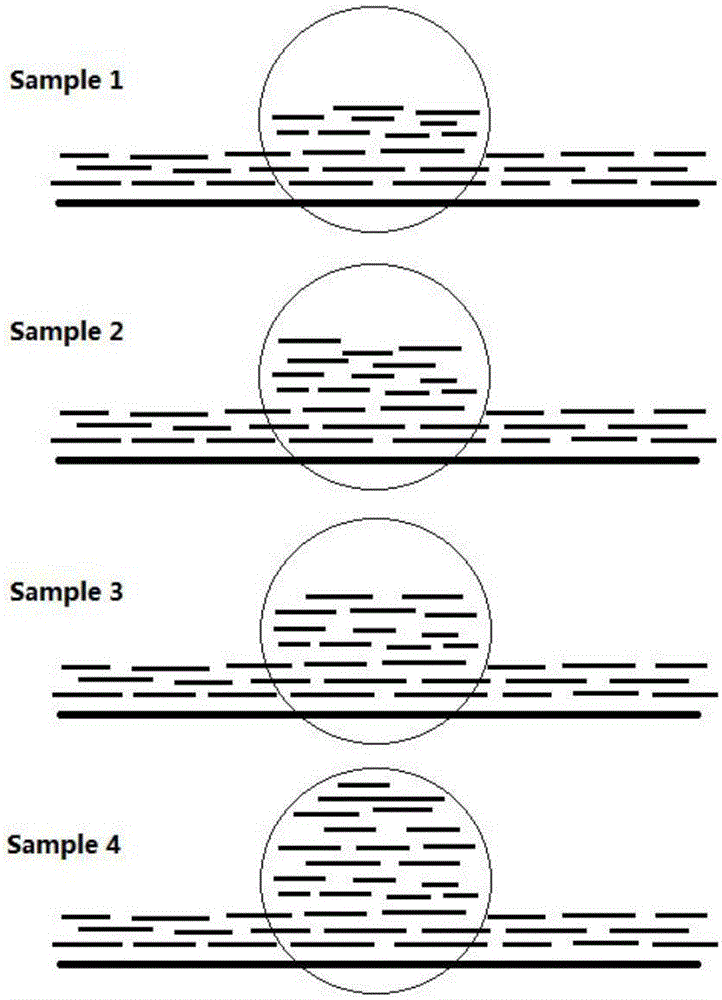
Method for genotyping forest populations on basis of gene CNV (copy number variation) sites
A technology of gene copy number and mutation site, applied in the field of molecular biology, can solve the problems of inability to accurately detect the genotype of CNV sites, difficulty in result analysis, application limitations, etc., so as to reduce the sequencing depth and reduce the algorithm complexity. , the effect of accurate typing results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] (1) Acquisition of raw materials: 435 individuals were collected from the natural distribution area of Populus tomentosa as research objects. After the genomic DNA of each individual strain was extracted by CTAB method, it was sent to Shanghai Bohao Biotechnology Co., Ltd. for sequencing. Two sequencing methods of pair-end of Illumina and mate-pair of 454 were used for sequencing to sequence Populus tomentosa individuals.
[0072] (2) Alignment: Using the above-mentioned alignment tools, software, and algorithms, compare the sequenced fragments (reads) obtained from each individual strain with the reference genome sequence, and remove PCR duplication, redundancy, and linker sequences introduced during the sequencing process.
[0073] (3) Statistics: use CNVnator software and its algorithm to count the relevant information of each potential CNV site area, such as the start-end coordinates of CNV, the length of CNV, type (deletion or duplication), read depth signal valu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com