Monomolecular gene sequencing method and device
A gene sequencing and single-molecule technology, applied in biochemical cleaning devices, biochemical equipment and methods, enzymology/microbiology devices, etc., can solve the problem of sequencing time, sequencing cost and unbiased sequencing that cannot meet the needs of biomedical testing, etc. problems, achieving fast sequencing time, improving signal-to-noise ratio, and wide application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0069] The parameters for processing data in this Example 1 are listed here:
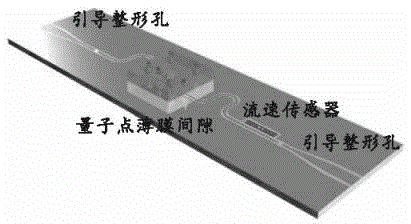
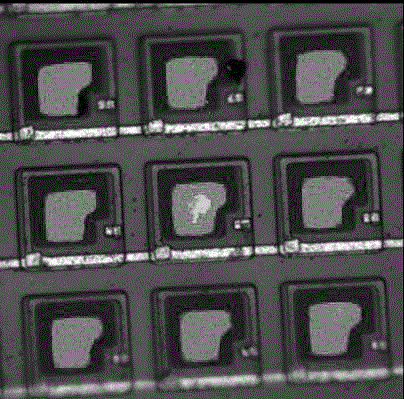
[0070] Step (1) The number of quantum dots is 2000-10000 per square micron, and the quantum dot spacing is greater than 0.5nm and less than 15nm;
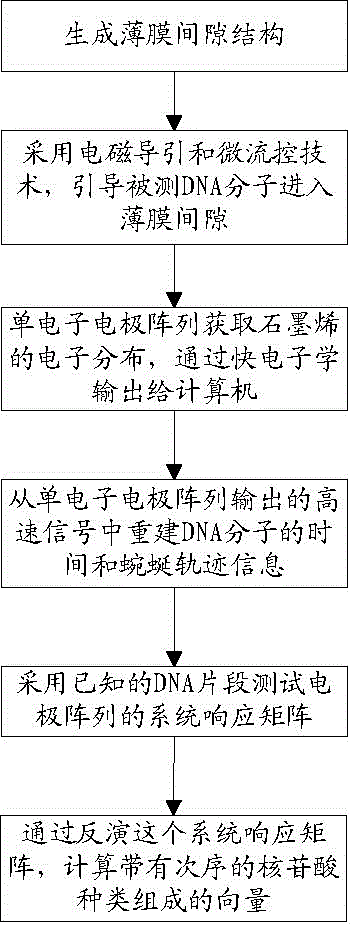
[0071] Step (2) use a strong electric field to drive a weak magnetic field to focus, and control the DNA strand to flow along the direction of the small hole at a speed of 5 microns to 5 mm per second;
[0072] Step (3) Using an ordinary desktop computer, the data throughput is between 1M / s and 8M / s;
[0073] Step (4) By controlling the flow rate, the total curvature of the DNA molecule is within 40 degrees;
[0074] Step (5) Obtain the system response matrix by means of experiment and simulation. After 5 independent tests, the error rate is required to be less than one millionth;
[0075] Step (6) uses an analytical large-scale linear equation solving method to directly draw the types of nucleotides at different positions.
example 2
[0077] The parameters for processing data in this application example 2 are listed here:
[0078] Step (1) The number of quantum dots is 200-50000 per square micron, and the quantum dot spacing is greater than 0.1nm and less than 15nm;
[0079] Step (2) use a strong electric field to drive a weak magnetic field to focus, and control the DNA strand to flow along the direction of the small hole at a speed of 50 microns to 10 mm per second;
[0080] Step (3) Use an ordinary smart phone as a data access carrier, which can be published online, and the data throughput is between 100k / s and 800k / s;
[0081] Step (4) By controlling the flow rate, the total curvature of the DNA molecule is within 30 degrees;
[0082] Step (5) Obtain the system response matrix by means of experiment and simulation. After 500 independent tests, the error rate is required to be less than 1 / 100,000;
[0083] Step (6) adopts an iterative large-scale linear equation solving method to approximate the types ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com