Primer and method for identifying leuconostoc mesenteroides subsp.mesenteroides and application of leuconostoc mesenteroides subsp.mesenteroides
A technology for identification of Leuconostoc enteritiformis and its identification method, which is applied in the field of primers for identifying Leuconostoc enteritidis subspecies, which can solve the time-consuming, cumbersome identification of Leuconostoc enteritidis subspecies, and the impossibility of subspecies identification. Level identification and other issues to achieve the effect of accelerated application and simple identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Primer design and validation for the identification of Leuconostoc enteritidis subspecies:
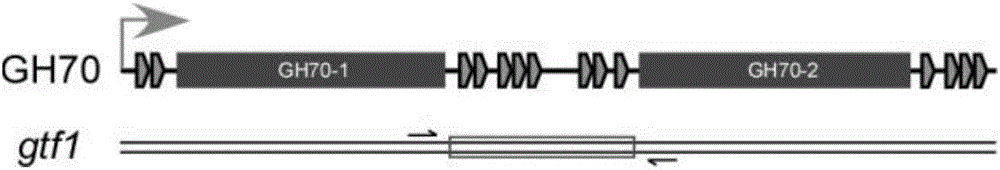
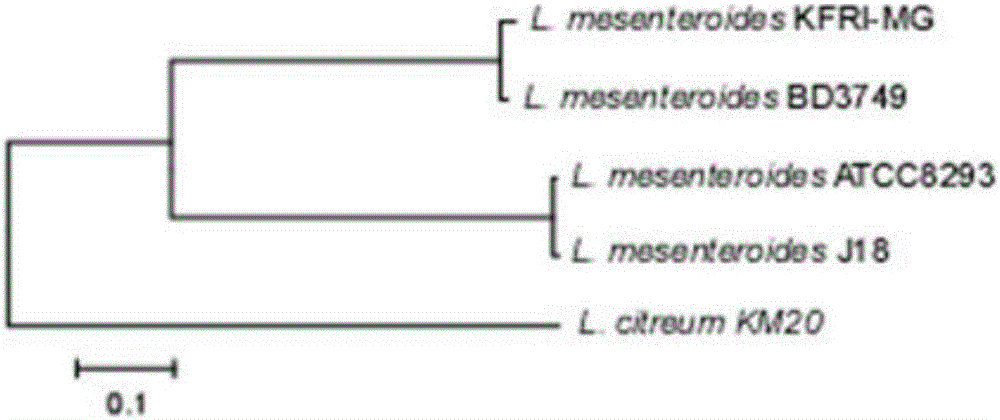
[0030] By comparing and analyzing the existing genome sequences of Leuconostoc enterococci (see attached table 1 for each strain and its genome sequence extraction code), a ubiquitous GH70 glycosyltransferase coding gene was identified and named gtf1. The gtf1 gene contains two highly conserved base sequences encoding the GH70 domain. Between these two highly conserved sequences, there is a highly variable sequence, which shows a high degree of polymorphism in different strains ( polymorphism).
[0031] Table 1 The complete genome information of Leuconostoc enterolis
[0032]
[0033] The schematic diagram of the gtf1 gene and its encoded product is attached figure 1 As shown, we designed primers in the highly conserved GH70 coding region to amplify the central highly variable region, and identified the strain by sequencing the amplified product.
[0034] According to the ...
Embodiment 2
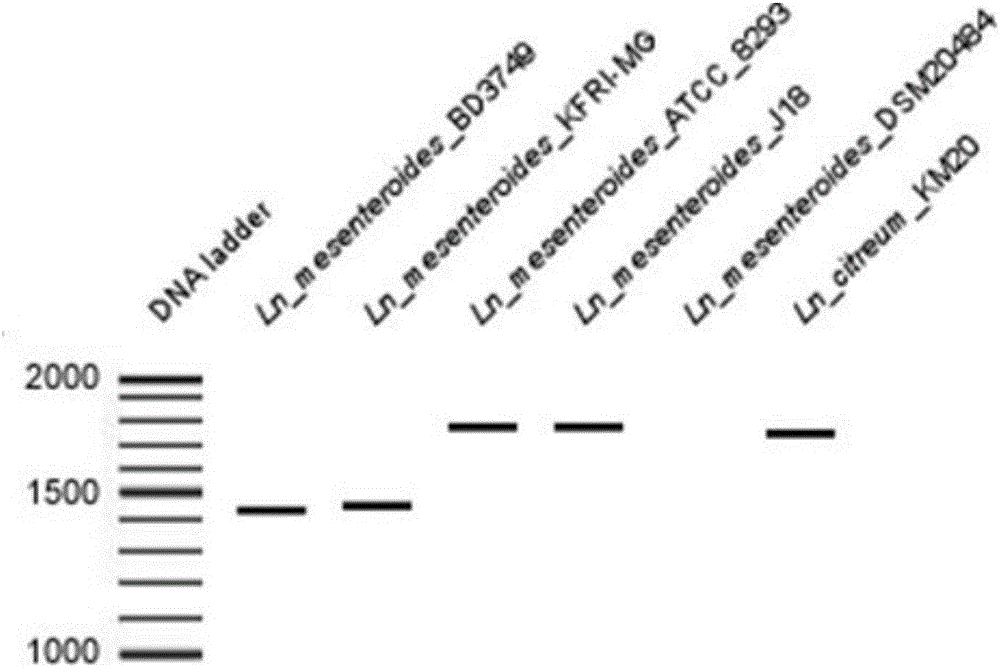
[0039] A method for identifying Leuconostoc enteritidis isolated from traditional foods at the level of Enteromenis subspecies, the method comprising the following steps:
[0040] (1) Taking Leuconostoc enterococci BD1710 isolated from traditional food in our laboratory as the test bacteria, the genomic DNA of the test strains was extracted, and the obtained DNA was used as a template for PCR amplification.
[0041] (2) Using the genomic DNA to be tested extracted from the above strains as a template, the PCR amplification reaction system is as follows: 2 μl of 2ng / μl DNA template, 1 μl of 0.5 μmol / L primer, 2 μl of 0.2mmol / L dNTP, 1.5mmol / L MgSO4 Add 1 μl, 2 μl of 10×PCR reaction buffer, 0.5 μl of 5U / μl rTaq DNA polymerase to 20 μl.
[0042] (3) Reaction parameters of PCR amplification: pre-denaturation at 95°C for 10 min; denaturation at 95°C for 30 s, annealing at 48°C for 30 s, extension at 72°C for 2 min, and 35 cycles; extension at 72°C for 15 min, and storage at 4°C.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com