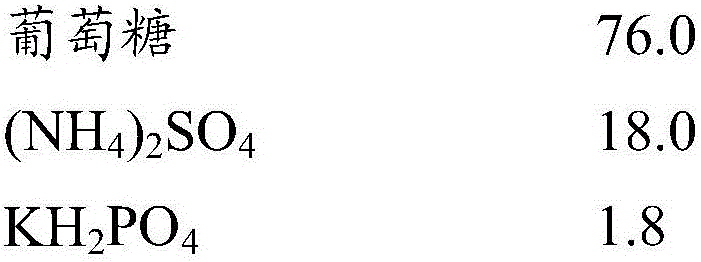Method For Producing An L-Amino Acid Using A Bacterium Of The Family Enterobacteriaceae Having An Attenuated Expression Of A gshA Gene
A technology of amino acids and Enterobacteriaceae, applied in biochemical equipment and methods, using vectors to introduce foreign genetic material, recombinant DNA technology, etc., can solve problems such as weakening the influence of gshA gene
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0174] Example 1: Construction of an E. coli strain with an inactivated gshA gene
[0175] 1.1 Construction of Escherichia coli MG1655ΔgshA strain
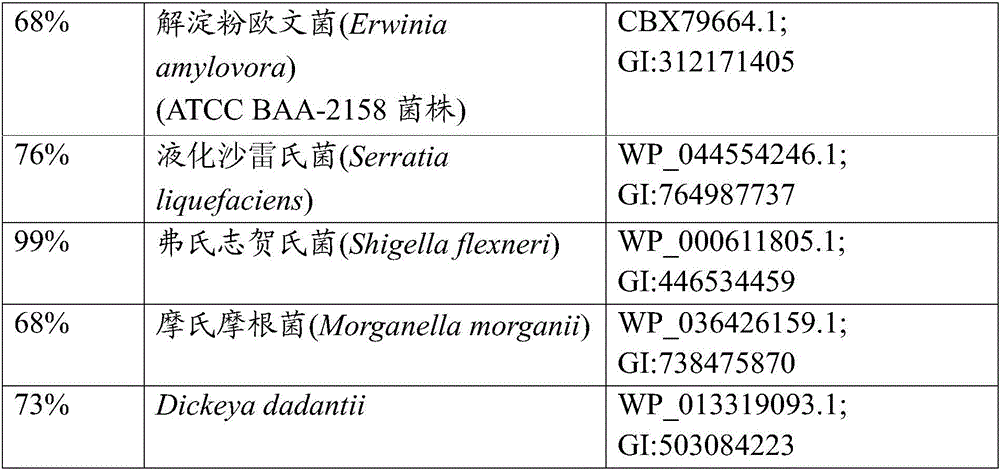
[0176] By a method originally developed by Datsenko K.A. and Wanner B.L. called "λRed / ET-mediated integration" (Datsenko K.A. and Wanner B.L., Proc.Natl.Acad.Sci.USA, 2000,97(12):6640- 6645) constructed an E. coli strain with an inactivated gshA gene. According to this method, PCR primers P1 (SEQ ID NO: 3) and P2 (SEQ ID NO: 4) were constructed, each of which was homologous to a region adjacent to the gshA gene at either end and adjacent to a region in the template plasmid conferring chlorine at the other end. Mycin resistance (Cm R ) regional homology of the cat gene. Plasmid pMIV-5JS (JP2008-99668, EP1942183 A1) was used as a template. The conditions of PCR are as follows: denaturation at 95°C for 3 minutes; profile of the first 2 cycles: 95°C for 1 minute, 34°C for 30 seconds, and 72°C for 1 minute; profile for the last 3...
Embodiment 2
[0181] Example 2: Construction of an L-valine-producing Escherichia coli strain with an inactivated gshA gene
[0182] In order to test the effect of the inactivation of the gshA gene on the production of L-valine, the Escherichia coli MG1655ΔgshA::Cm R The DNA fragment of the chromosome of the bacterial strain (Example 1.1) was transferred to a valine-producing Escherichia coli H81 strain (Miller J.H., Experiments in Molecular Genetics, Cold Spring Harbor Lab. Press, Plainview, NY, 1972) by P1 transduction to obtain large intestine Bacillus H81ΔgshA::Cm R strain. H81 bacterial strain (EP1239041A2) was preserved in Russian National Industrial Microorganism Collection Center (VKPM; FGUP GosNII Genetika, Russia, zip code 117545, Moscow (RussianFederation, 117545Moscow, 1) with preservation number VKPM B-8066 on January 30, 2001 st Dorozhny proezd,1)), which was then transferred to an international deposit on February 1, 2002 under the terms of the Budapest Treaty. E. coli H8...
Embodiment 3
[0183] Example 3: by E. coli H81ΔgshA::Cm R Strains for production of L-valine
[0184]The modified Escherichia coli H81ΔgshA::Cm R and the control Escherichia coli H81 strain were cultured in 2 mL LB medium at 34°C for 18 hours. Then, 0.2 mL of the obtained culture was each inoculated into 2 mL of fermentation medium in a 20×200-mm test tube and cultured on a rotary shaker (250 rpm) at 34° C. for 72 hours until glucose was consumed.
[0185] The composition (g / L) of fermentation medium is as follows:
[0186]
[0187]
[0188] The fermentation medium was sterilized at 116°C for 30 minutes. Glucose and CaCO 3 Sterilize separately as follows: Glucose was sterilized at 110 °C for 30 min while CaCO 3 Sterilize at 116°C for 30 minutes. The pH was adjusted to 7.0 by adding 6M KOH before sterilization.
[0189] After culturing, the amount of L-valine accumulated in the medium was determined by thin layer chromatography (TLC). Merck silica gel 60-coated 10 x 20-cm TLC p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com