ZNA (zip nucleic acid) primer based STR (short tandem repeat) typing kit
A kit and forward primer technology, which is applied in the field of forensic DNA testing and analysis, can solve problems such as poor typing effect, inability to meet testing requirements at the same time, and unsuitability for the Chinese population.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
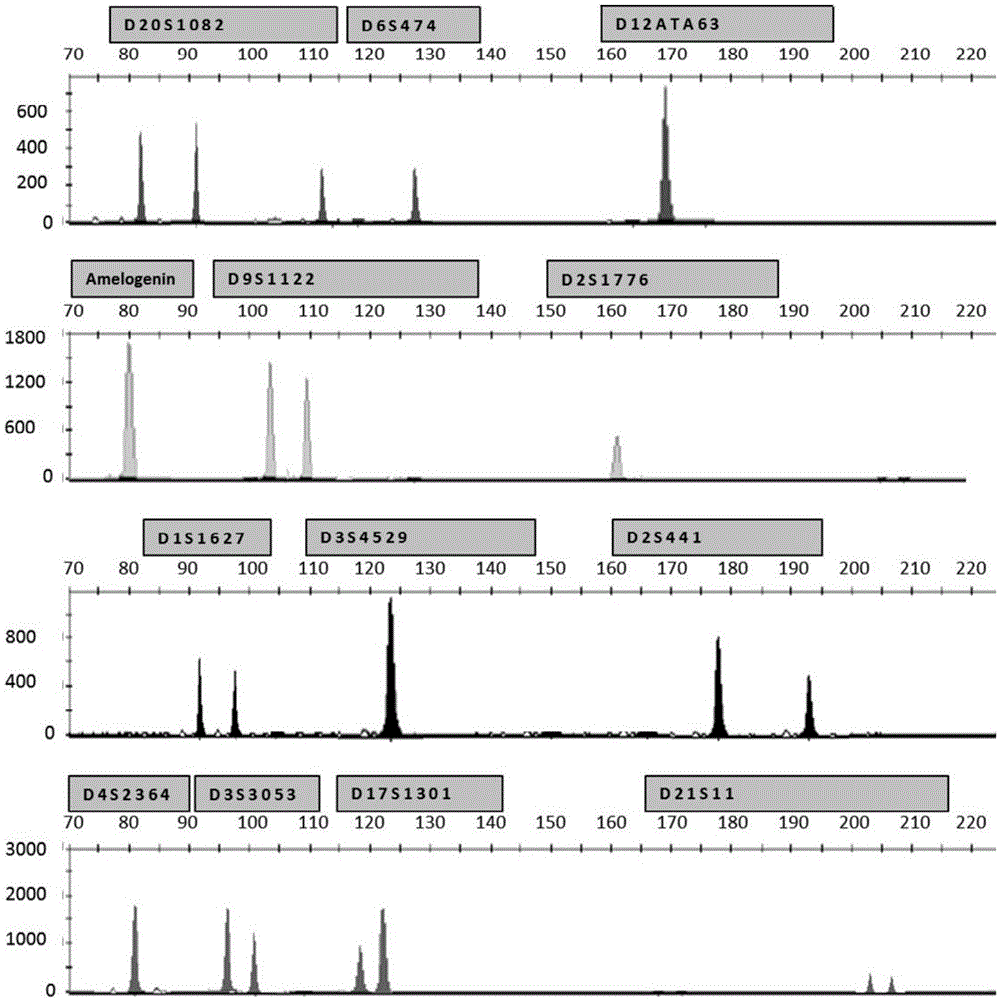
[0153] Example 1 Genotyping of 10 different volunteers
[0154] 1. Sample collection: The sample is blood from volunteers, 10 samples in total.
[0155] 2. Template DNA extraction: Template DNA was extracted from 10 different volunteers' blood samples by Chelex-100 method.
[0156] 3. PCR amplification:
[0157] (1), the ZNA primers used are:
[0158] A pair of ZNA primers used to amplify D20S1082 are: Forward primer: The nucleic acid sequence shown in SEQ ID NO: 1 is a ZNA sequence composed of a Z unit coupled to the 9th and 15th nucleotides from the 5' end, Reverse primer: a ZNA sequence composed of a Z unit coupled to the 5th, 11th and 15th nucleotides from the 5' end of the nucleic acid sequence shown in SEQ ID NO: 2;
[0159] A pair of ZNA primers used to amplify D6S474 are: Forward primer: The nucleic acid sequence shown in SEQ ID NO: 3 is a ZNA sequence composed of a Z unit coupled to the 5th and 21st nucleotides from the 5' end, Reverse primer: the nucleic acid seq...
experiment example 2
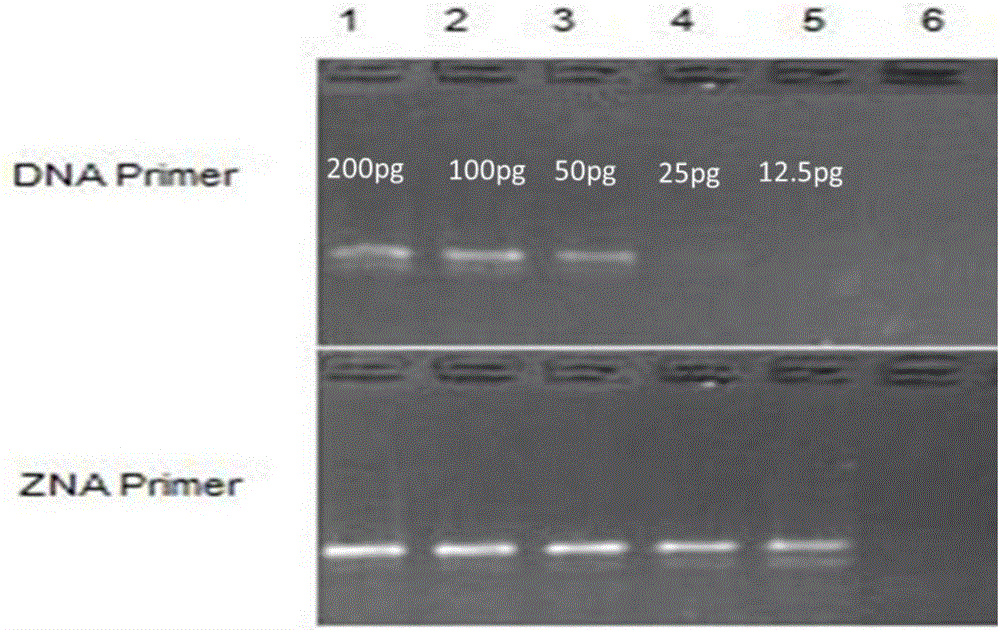
[0186] Experimental example 2 template concentration
[0187] Taking the D20S1082 locus as an example, the blood DNA of a volunteer in Example 1 was used as a template, and its ZNA primer and DNA primer were used respectively, and the template amounts were 200, 100, 50, 25, 12.5, and 6.25 pg respectively. PCR amplification, PCR amplification system and amplification program are with embodiment 1, amplified product carries out agarose gel electrophoresis, the result is as follows figure 2 shown. Under the same amplification conditions, the concentration of the amplification product obtained by ZNA primers was significantly higher than that of DNA primers, and ZNA primers could still see clear amplification bands when the template amount was 12.5pg, while DNA primers, under the same conditions, The amplified bands with a template amount of 50 pg began to be blurred. The template concentration of the remaining loci was the same as that of the D20S1082 locus.
[0188] case:
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com