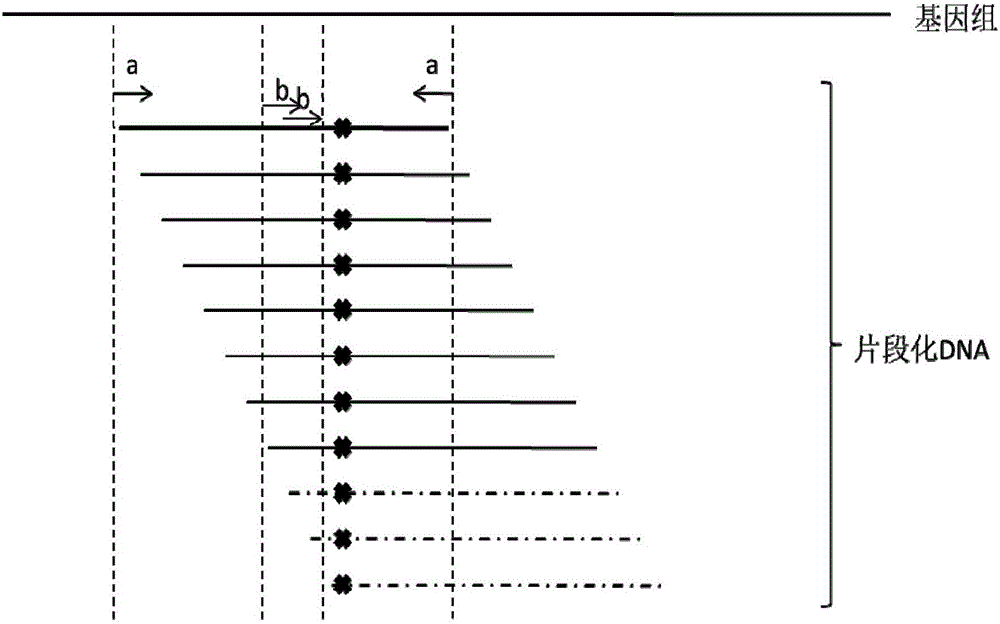
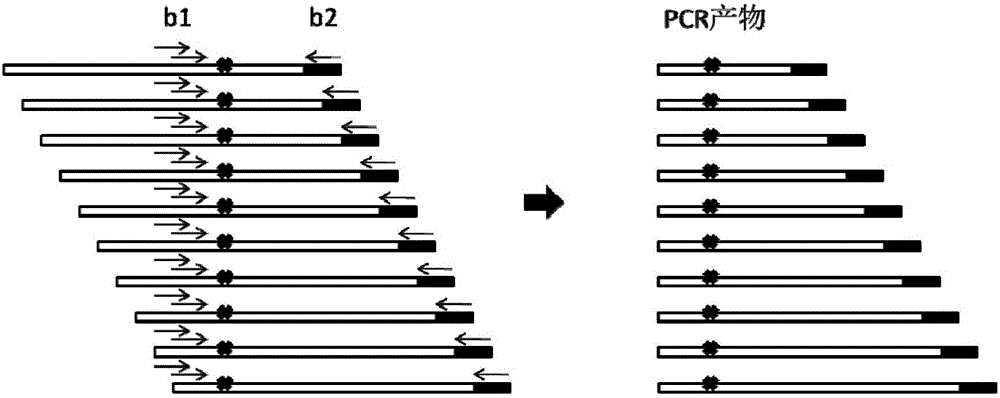
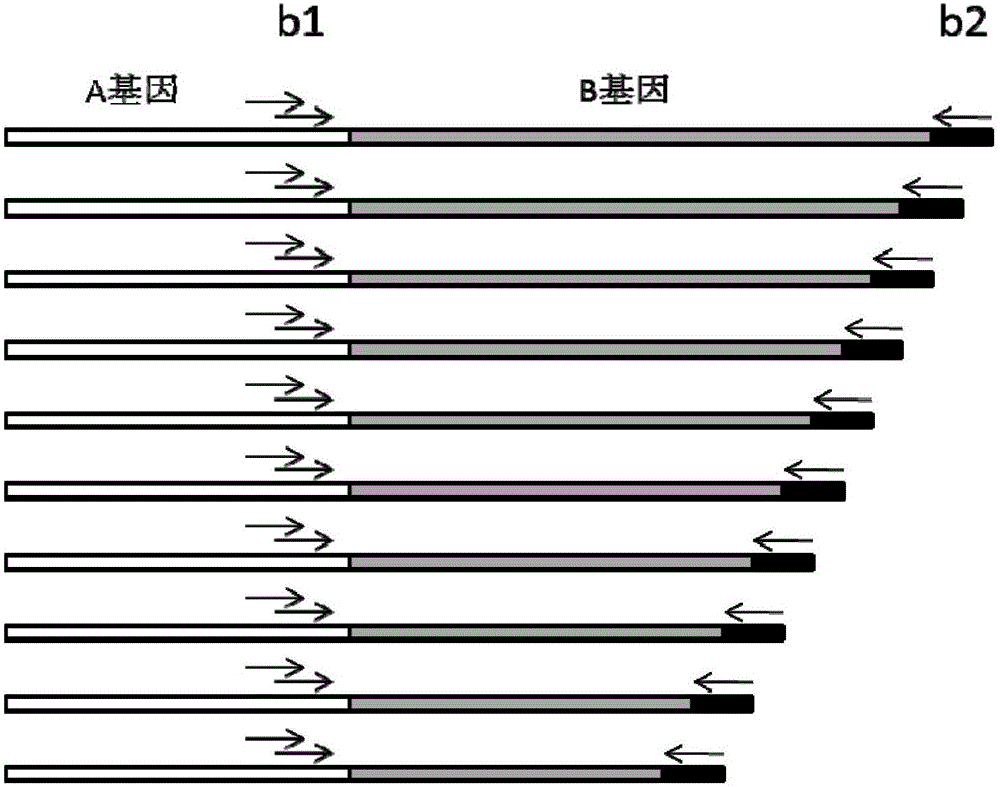
Primer composition for detecting fragmentized DNA target area and application thereof
A technology of primer composition and target region, which is applied in the field of primer composition for detection of fragmented DNA target region, can solve the problems of low mixing ratio, need for improvement, detection method is difficult to achieve such a high detection sensitivity, etc., to improve the scope of application , The sequencing results are accurate and reliable, and the effect of improving the scope of adaptation and the amount of effective templates
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0133] 1. Connector design
[0134] Prelib-ADT-S: CAAGCAGAAGACGGCATACGAGATIIIIIIIGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (I is the index area),
[0135] Prelib-ADT-AS: pGATCGGAAGAGC,
[0136] The above sequences need to anneal into double strands.
[0137] The structure of the product after connection:
[0138] Top: 5'-CAAGCAGAAGACGGCATACGAGATIIIIIIIGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT---AGATCGGAAGAGC-3',
[0139] Bottom: 5'-CAAGCAGAAGACGGCATACGAGATIIIIIIIGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT---AGATCGGAAGAGC-3',
[0140] Co-extension-specific primers were designed for exon 19 of EGFR.
[0141] The sequence of exon 19 of EGFR is as follows:
[0142] GGACTCTGGATCCCAGAAGGTGAGAAAGTTAAAATTCCCGTCGCTATCAAGGAATTAAGAGAAGCAACATCTCCGAAAGCCAACAAGGAAATCCTCGAT (SEQ ID NO: 11),
[0143] The sequences of exon 19 of EGFR and its upstream and downstream intron regions are as follows:
[0144] CAGCCCCCAGCAATATCAGCCTTAGGTGCGGCTCCACAGCCCCAGTGTCCCTCACCTTCGGGGTGCATCGCTGGTAACATCCACCCAGATCACTGGGCAGCATGTG...
Embodiment 2
[0221] 1. Connector design
[0222] Prelib-ADT-S: CAAGCAGAAGACGGCATACGAGATIIIIIIIGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (I is the index area),
[0223] Prelib-ADT-AS: pGATCGGAAGAGC,
[0224] The above sequences need to anneal into double strands.
[0225] The structure of the product after connection:
[0226] Top: 5'-CAAGCAGAAGACGGCATACGAGATIIIIIIIGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT---AGATCGGAAGAGC-3',
[0227] Bottom: 5'-CAAGCAGAAGACGGCATACGAGATIIIIIIIGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT---AGATCGGAAGAGC-3',
[0228] Direct extension-specific primers were designed for the T790M point mutation in exon 20 of EGFR.
[0229] The sequence of exon 20 of EGFR is as follows:
[0230] GAAGCCTACGTGATGGCCAGCGTGGACAACCCCCACGTGTGCCGCCTGCTGGGCATCTGCCTCACCTCCACCGTGCAGCTCATCACGCAGCTCATGCCTCCTGGCTGCCTCCTGGACTATGTCCGGGAACACAAAGACAATATTGGCTCCCAGTACCTGCTCAACTGGTGTGTGCAGATCGCAAAG (SEQ ID NO: 13),
[0231] The sequences of exon 20 of EGFR and its upstream and downstream intron regions are as follow...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com