Use of human TNFRSF12A gene and related drugs
A gene and drug technology, applied in the application of human TNFRSF12A gene and related drugs, can solve the problem of few reports of TNFRSF12A
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1 Preparation of RNAi lentivirus for human TNFRSF12A gene
[0081] 1. Screening for effective siRNA targets against human TNFRSF12A gene
[0082] Retrieve TNFRSF12A gene information from Genbank; design effective siRNA targets for TNFRSF12A gene. Table 1 lists the effective siRNA target sequences for the TNFRSF12A gene.
[0083] Table 1 is targeted at the siRNA target sequence of human TNFRSF12A gene
[0084]
[0085] 2. Preparation of lentiviral vector
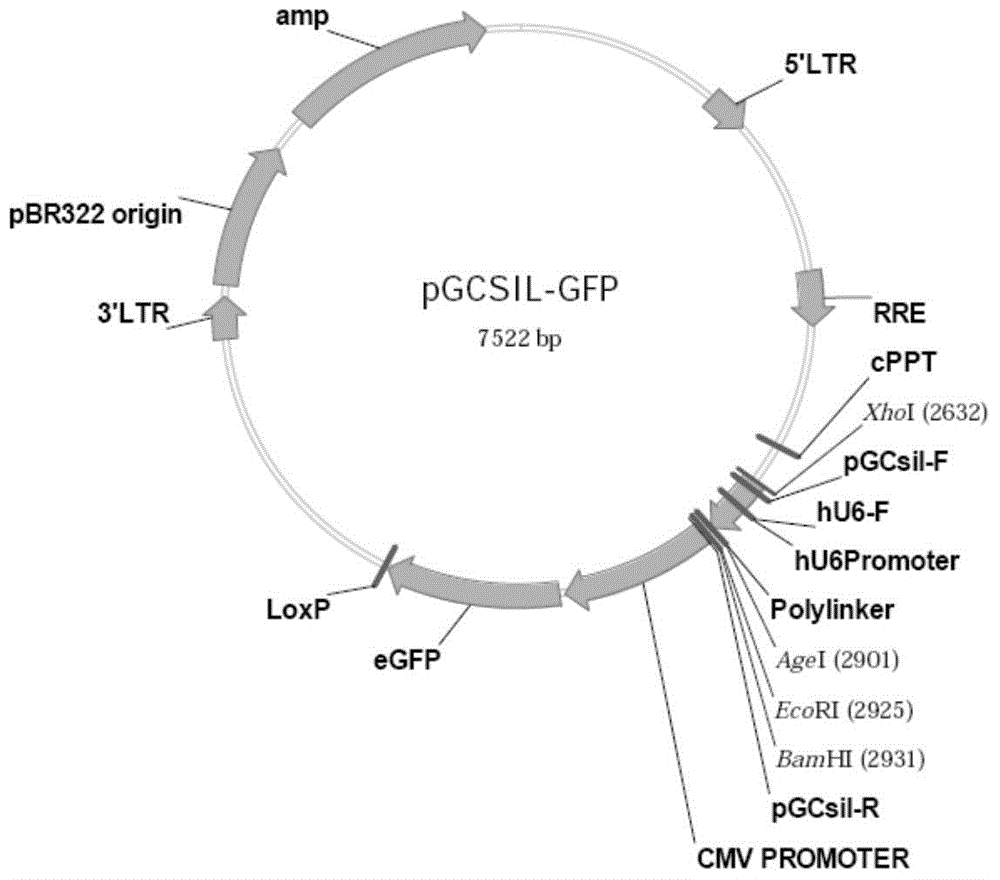
[0086] Aiming at the siRNA target (taking SEQIDNO: 1 as an example) synthetic double-stranded DNA Oligo sequence (table 2) containing AgeI and EcoRI restriction endonuclease at both ends; Acting on the pGCSIL-GFP vector ( Provided by Shanghai Jikai Gene Chemical Technology Co., Ltd., figure 1 ), to linearize it, and identify the digested fragments by agarose gel electrophoresis.
[0087] Table 2 Double-stranded DNA Oligo with sticky ends containing AgeI and EcoRI restriction sites at both ends
[0088]...
Embodiment 2
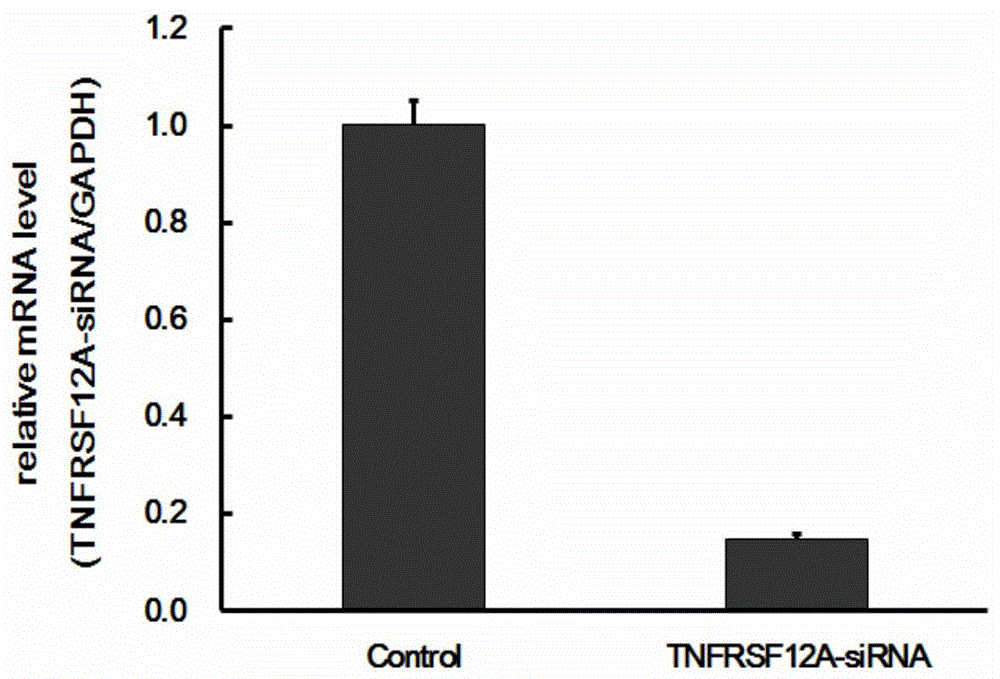
[0107] Example 2 Real-time fluorescent quantitative RT-PCR method to detect the silencing efficiency of TNFRSF12A gene
[0108] Human liver cancer SMMC-7721 cells in the logarithmic growth phase were digested with trypsin to make a cell suspension (the number of cells was about 5×10 4 / ml) were inoculated in a 6-well plate and cultured until the cell confluency reached about 30%. According to the multiplicity of infection (MOI, SMMC-7721:10) value, an appropriate amount of the virus prepared in Example 1 was added, the culture medium was replaced after 24 hours of cultivation, and the cells were collected after the infection time reached 4 days. Total RNA was extracted according to the instruction manual of Invitrogen's Trizol. According to the M-MLV instruction manual of Promega Company, RNA was reverse-transcribed to obtain cDNA (see Table 7 for the reverse transcription reaction system, react at 42° C. for 1 h, and then bathe in a water bath at 70° C. for 10 min to inactiv...
Embodiment 3
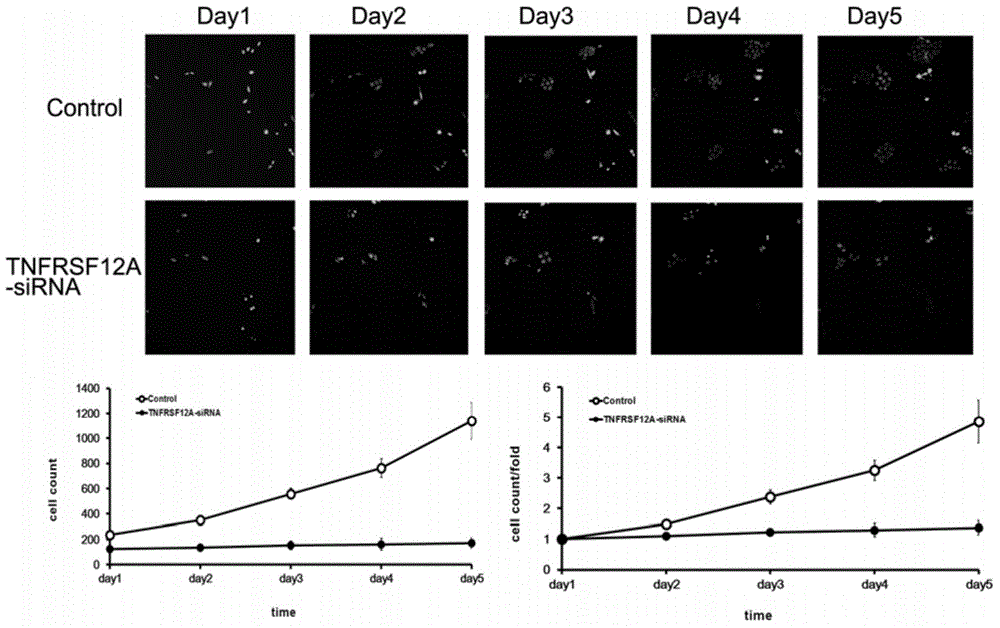
[0115] Example 3 Detection of proliferation ability of tumor cells infected with TNFRSF12A-siRNA lentivirus
[0116] Human liver cancer SMMC-7721 in the logarithmic growth phase was digested with trypsin to make a cell suspension (the number of cells was about 5×10 4 / ml) were inoculated in a 6-well plate and cultured until the cell confluency reached about 30%. According to the multiplicity of infection (MOI, SMMC-7721:10), an appropriate amount of virus was added, and the culture medium was replaced after 24 hours of culture. After the infection time reached 4 days, the cells of each experimental group in the logarithmic growth phase were collected. The complete medium was resuspended into a cell suspension (2×10 4 / ml), inoculate a 96-well plate at a cell density of about 2000 / well. 5 replicate wells in each group, 100 μl per well. After laying the board, place at 37°C, 5% CO 2 Incubator cultivation. From the second day after plating, the plate was detected and read on...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com