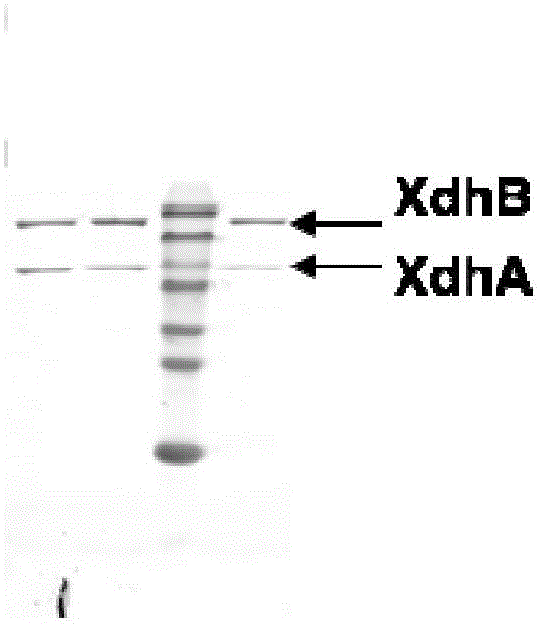Xanthine dehydrogenase, and coding gene thereof and application
A coding gene and coding technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve the problems such as no commercial microbial source XDH report, achieve a wide range of pH tolerance and temperature tolerance, reduce costs , the effect of reducing the dosage of enzymes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
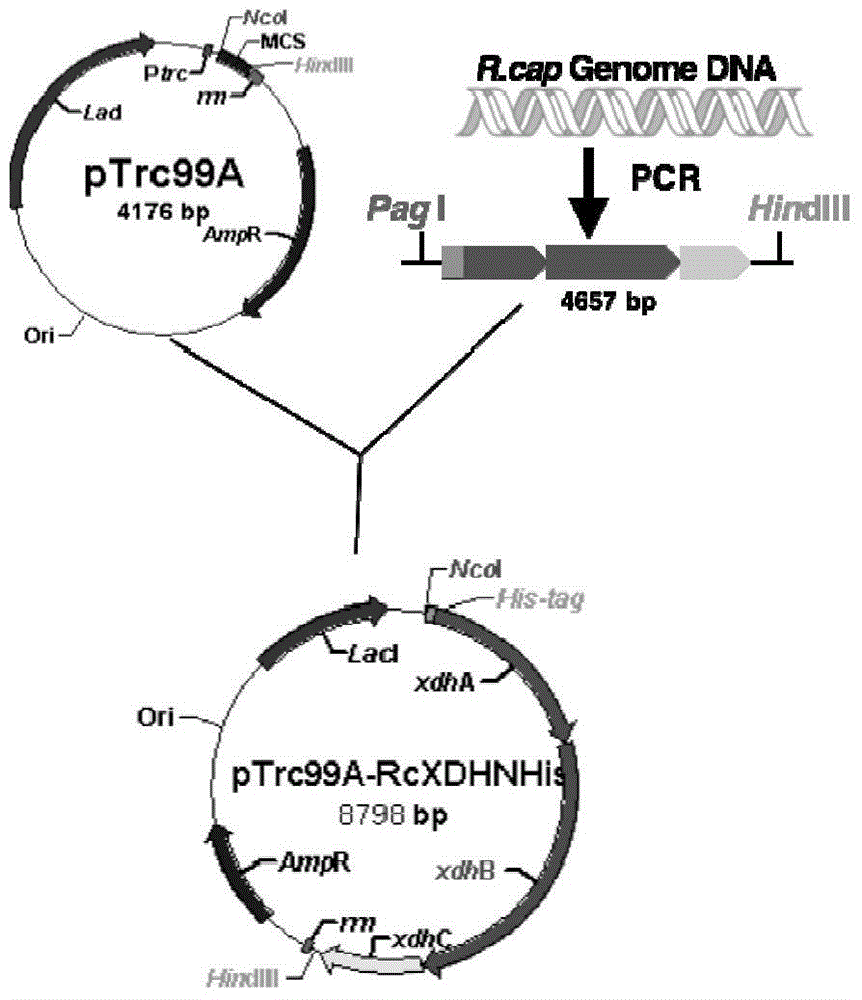
[0044] Embodiment 1, the construction of the recombinant plasmid pTrc99A-RcXDHNHis containing the rhodobacter capsularis xanthine dehydrogenase gene
[0045] 1. Extract the genomic DNA of Rhodobacter capsulatus with the preservation number CGMCC1.3366.
[0046] 2. Design and synthesize the following primers
[0047] Upstream primer: 5'-GGC TCATGA TGCATCATCATCACCATCACCATATGGAAATTGCGTTTCTTTCTCAATG-3' (SEQ ID No. 1)
[0048] (The underlined sequence is the recognition site for PagI digestion, and the italic part is the coding sequence of the 6xHistag purification tag introduced for the convenience of purification)
[0049] Downstream primer: 5'-GCC AAGCTT ATCACCCGGTCTGTCCTTCC-3' (SEQ ID No. 2)
[0050] (The underlined sequence is the recognition site for HindIII digestion)
[0051] 3. Using the genomic DNA of rhodobacter capsularis extracted in step 1 as a template, and using the upstream primer and the downstream primer as primers, PCR amplification is carried out to obta...
Embodiment 2
[0055] Embodiment 2, the purification of recombinant xanthine dehydrogenase
[0056] 1. Transform pTrc99A-RcXDHNHis into Escherichia coli DH5α to obtain recombinant bacteria, pick a single colony of recombinant bacteria, inoculate in 5ml LB liquid medium containing 100μg / ml ampicillin, and culture overnight at 37°C and 220r / min. Transfer the overnight cultured bacterial solution to 500ml LB liquid medium containing 100μg / mL ampicillin according to the inoculum size of 1%, and cultivate it at 37°C and 220r / min until the bacterial solution is cultivated to OD 600 0.6, add IPTG inducer with a final concentration of 1mmol / L, and continue to induce culture for 16h under the same conditions.
[0057] 2. Centrifuge the bacterial solution obtained in step 1 at 9000r / min and 4°C, collect the precipitate, and obtain the engineered bacteria induced by expression. After the precipitate is washed once with 50mmol / L phosphate buffer (pH7.5), it is resuspended in 100ml of the same In the bu...
Embodiment 3
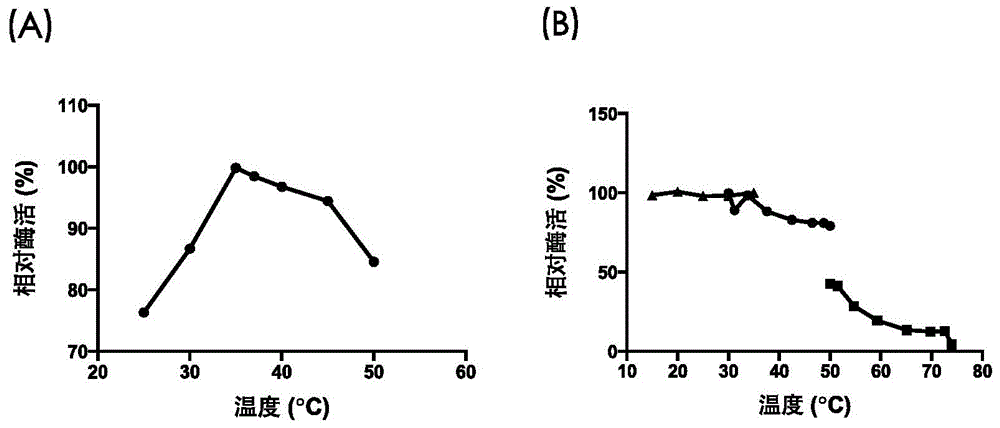
[0065] Embodiment 3, the characterization of recombinant xanthine dehydrogenase
[0066] One, the assay method of xanthine dehydrogenase enzymatic activity
[0067] The 2mL reaction system is shown in Table 1.
[0068] The 2mL reaction system composition of table 1 xanthine dehydrogenase activity assay
[0069]
[0070] Mix the remaining reagents in the 2mL reaction system in Table 1 except the aqueous solution of recombinant xanthine dehydrogenase prepared in Example 2 and place it in a 35°C water bath for 5 minutes, then add 100 μL of the recombinant xanthine dehydrogenase prepared in Example 2 An aqueous enzyme solution initiates the reaction. Record the OD within 3-5min of the reaction while reacting at 35°C 295 Absorbance value change, calculate the rate of change of absorbance with time in the initial linear part of the reaction curve (ΔOD 295 / min), the rate of change of absorbance with time (ΔOD 295 / min) reflects the rate at which the recombinant xanthine dehy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com