Pyrrolo-triazinone derivatives
A compound and solvate technology, applied in the field of pyrrolotriazinone derivatives, can solve problems such as inability to repair DNA fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] 1-[3-(4-Cyclopentylcarbonyl-1-piperazine-1-formyl)-benzyl]-3H-pyrrolo[1,2-d][1,2,4]triazine-4 -Synthesis of ketone (1)
[0082]
[0083] 3-(4-Oxo-3,4-dihydropyrrolo[1,2-d][1,2,4]triazin-1-yl-methyl)benzoic acid (2.0 g, 7.43 mmol) , cyclopentyl-piperazin-1-yl-methanone (1.4g, 7.68mmol), O-benzotriazole-N, N, N', N'-tetramethylurea tetrafluoroborate ( TBTU) (3.6g, 11.21mmol) and N,N-diisopropylethylamine (2.9g, 22.44mmol) were added to dimethylformamide (20ml) and stirred for 3 hours. Water (100ml) was added, and the pH of the reaction solution was adjusted with 1N HCl under stirring. A large amount of solids were precipitated, and hydrochloric acid was continued to be added dropwise until no solids precipitated. Filter, wash the filter cake successively with a large amount of water and a small amount of ethanol, and dry in vacuum to obtain 1-[3-(4-cyclopentylcarbonyl-1-piperazine-1-formyl)-benzyl]-3H-pyrrolo [1,2-d][1,2,4]triazin-4-one (1) white solid (3.0 g, 93.0%...
Embodiment 2
[0088] 1-[3-(4-Cyclopropylcarbonyl-1-piperazine-1-formyl)-4-fluorophenyl]-3H-pyrrolo[1,2-d][1,2,4]tri Synthesis of oxazin-4-one (4)
[0089]
[0090] 2-Fluoro-5-(4-oxo-3,4-dihydropyrrolo[1,2-d][1,2,4]triazin-1-yl-methyl)benzoic acid (1.5g , 5.22mmol), cyclopropyl-piperazin-1-yl-methanone (0.48g, 5.58mmol), O-benzotriazole-N, N, N', N'-tetramethylurea tetrafluoro Borate ester (TBTU) (2.5g, 7.79mmol) and N,N-diisopropylethylamine (2.0g, 15.50mmol) were added to dimethylformamide (15ml), and stirred for 2-3 hours. Water (100ml) was added, and the pH of the reaction solution was adjusted with 1N HCl under stirring. A large amount of solids were precipitated, and hydrochloric acid was continued to be added dropwise until no solids precipitated. Filtration, the filter cake was successively rinsed with a large amount of water and a small amount of ethanol, and dried in vacuum to obtain 1-[3-(4-cyclopentylcarbonyl-1-piperazine-1-formyl)-4-fluorophenyl]-3H - pyrrolo[1,2-d][1,2,4]...
Embodiment 3
[0095] To assess the inhibitory effect of compounds, the IC was determined using the following assay 50 value.
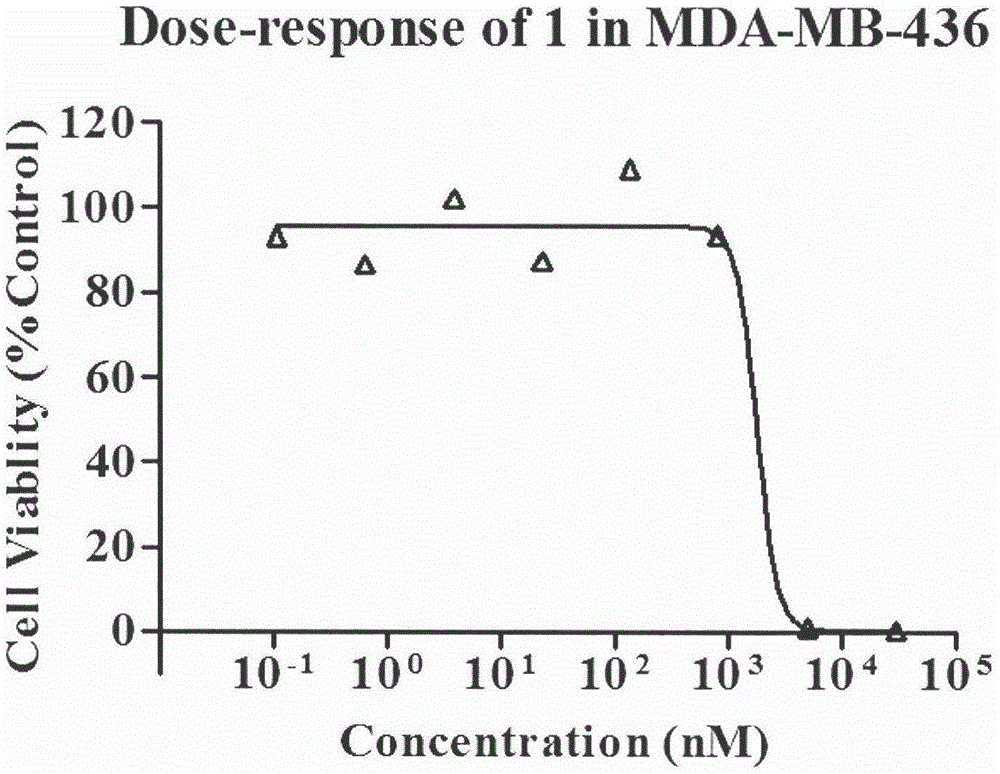
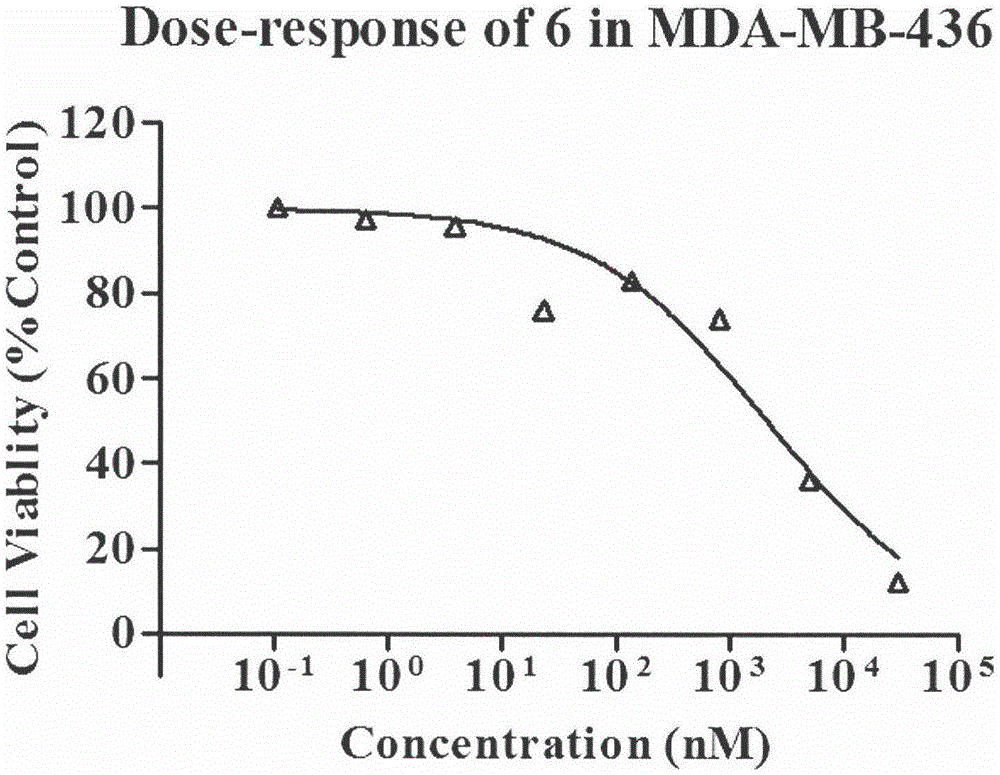
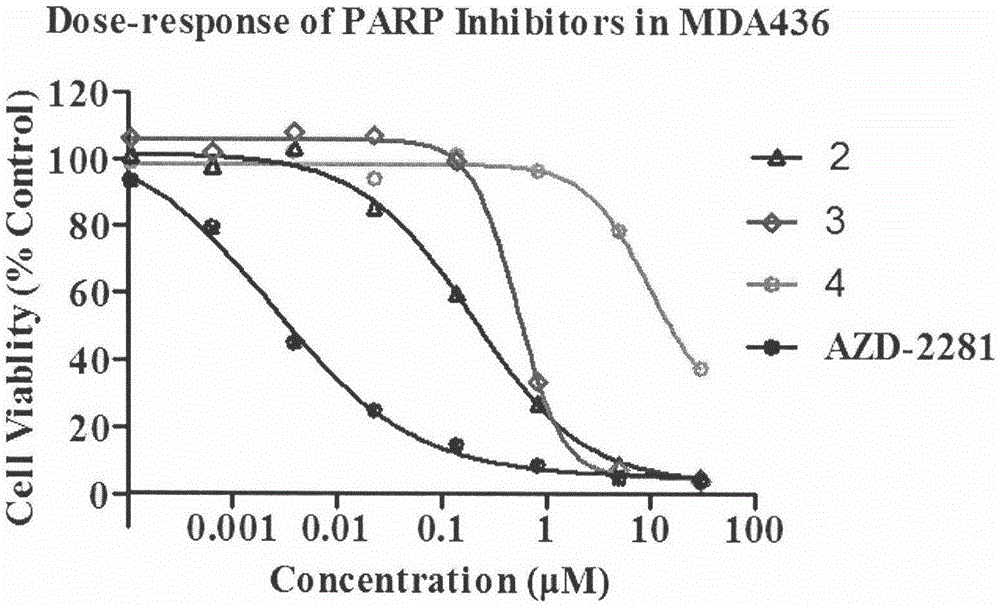
[0096] Using the Alarmblue method to determine the IC of the test sample and positive control AZD2281 in MDA-MB-436 tumor cells 50 , 9 concentration gradients (including 0 concentration) were set up for each compound, and 2 replicate wells were used.
[0097] MDA436 cells were cultured in MEM complete medium (containing 10% fetal bovine serum, 100 U·mL -1 Penicillin, 100 μg mL -1 Streptomycin), at 37°C, 5% CO 2 , 2 to 3 generations under 95% air culture conditions.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com