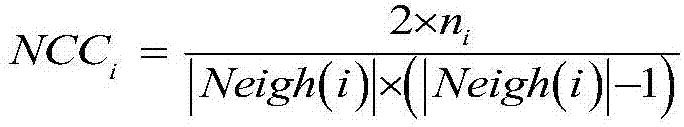A method for identifying protein complexes based on BSO optimization
A protein complex and identification method technology, applied in the field of protein complex identification based on BSO optimization, can solve unsustainable, time-consuming, expensive and other problems, and achieve significant bioaccumulation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] Below in conjunction with specific embodiment, the present invention is further described:
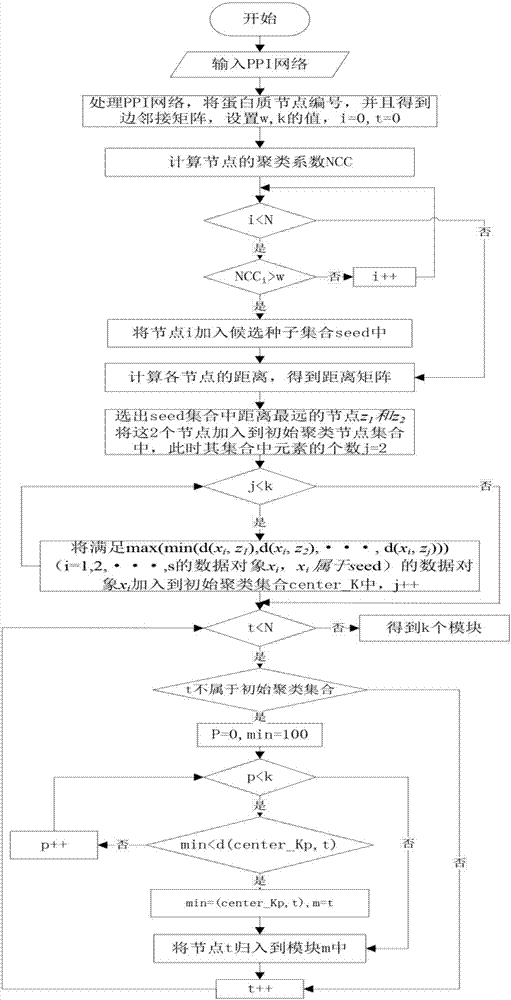
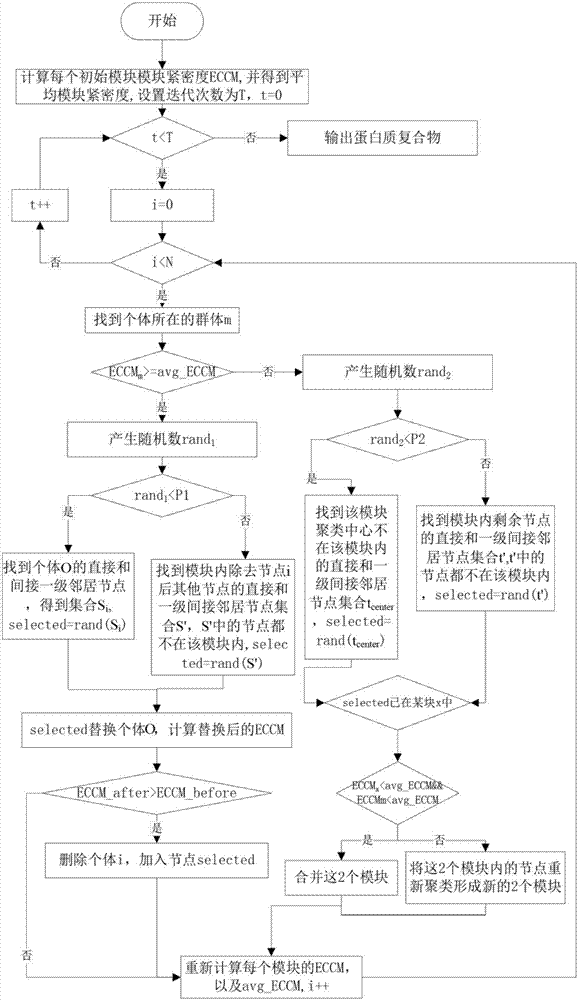
[0042] Such as figure 1 In the first step of the protein complex identification method based on BSO optimization, the idea of k-means algorithm is used to select k initial cluster centers among all nodes, and the distances from other nodes to each cluster center are compared. It is grouped into the module where the nearest cluster centers are located. Its specific implementation steps include:
[0043] (1) Selection of candidate seeds
[0044] The PPI network is abstracted as a network connectivity graph G=(V, E) formed by multiple protein nodes and the interactions between nodes, where V is the set of protein nodes, and E is the set of interacting edges between protein nodes. Each protein complex starts from a given node and is clustered to obtain a module. For the convenience of explanation, the definition of the clustering coefficient of the direct neighbor set and the no...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com