D-lactic dehydrogenase mutant and application thereof
A technology of phenyllactic acid and amino acid, applied in the field of D-lactate dehydrogenase mutants, which can solve the problems of complicated separation and purification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1. Preparation of D-lactate dehydrogenase mutant
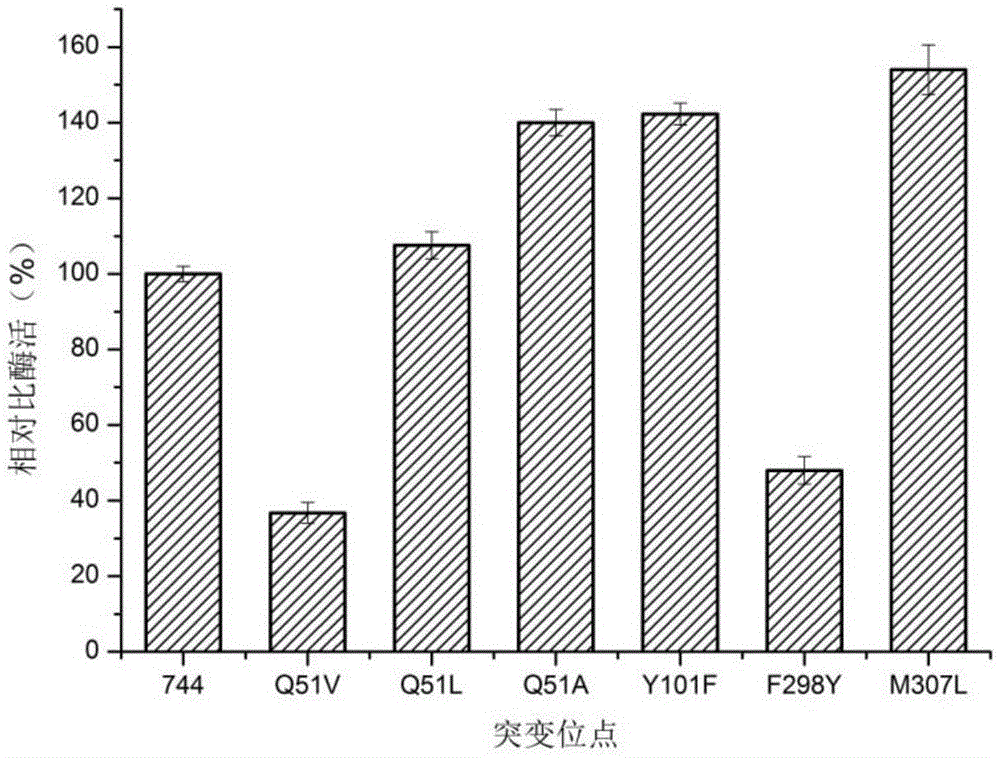
[0038] According to the simulated three-dimensional structure of the DLDH744 protein containing the substrates PPA and NADH, the glutamine (Q) close to the C3 end of the PPA substrate was mutated into smaller hydrophobic amino acids alanine (A) and valine, respectively. Acid (V), leucine (L), tyrosine (Y) at position 101 was mutated to phenylalanine (F), and phenylalanine (F) at position 298 was mutated to tyrosine (Y) ), 307-position methionine (M) was mutated to leucine (L), and D-lactate dehydrogenase mutants Q51V, Q51L, Q51A, Y101F, F298Y, and M307L mutants were constructed.
[0039] The amino acid sequence of wild-type D-lactate dehydrogenase is sequence 2, and the nucleotide sequence of its encoding gene is sequence 1.
[0040] The amino acid sequence of D-lactate dehydrogenase mutant Q51V is the sequence obtained by mutating Gln at position 51 of SEQ ID NO: 2 to Val; - Sequence obtained by mutating CA a...
Embodiment 2
[0064] Example 2. Determination of enzyme activity of D-lactate dehydrogenase mutants
[0065] Determination of enzyme activity of D-lactate dehydrogenase mutant In view of the fact that coenzyme NADH has a maximum absorption peak at 340nm, the enzyme activity is defined by the decreasing speed of the absorbance value of NADH at 340nm, as follows:
[0066] Phenylpyruvic acid solution: use phenylpyruvic acid and 100mmol / L phosphate buffer (recipe: weigh 79gNaCl, 2gKCl, 2.4gKH 2 PO 4 and 1.8gK 2 HPO 4 , dissolve in 800ml of distilled water, adjust the pH of the solution to 7.4 with HCl, and finally add distilled water to dilute to 1L. Store in a 4°C refrigerator. ) mixing, the obtained solution, and the concentration of phenylpyruvate is 200mM;
[0067] NADH solution: mix with NADH and 100mmol / L phosphate buffer to obtain a solution, and the concentration of NADH is NADH.
[0068] 200 μl reaction system: 10 μL phenylpyruvate solution, 10 μL NADH solution and appropriate en...
Embodiment 3
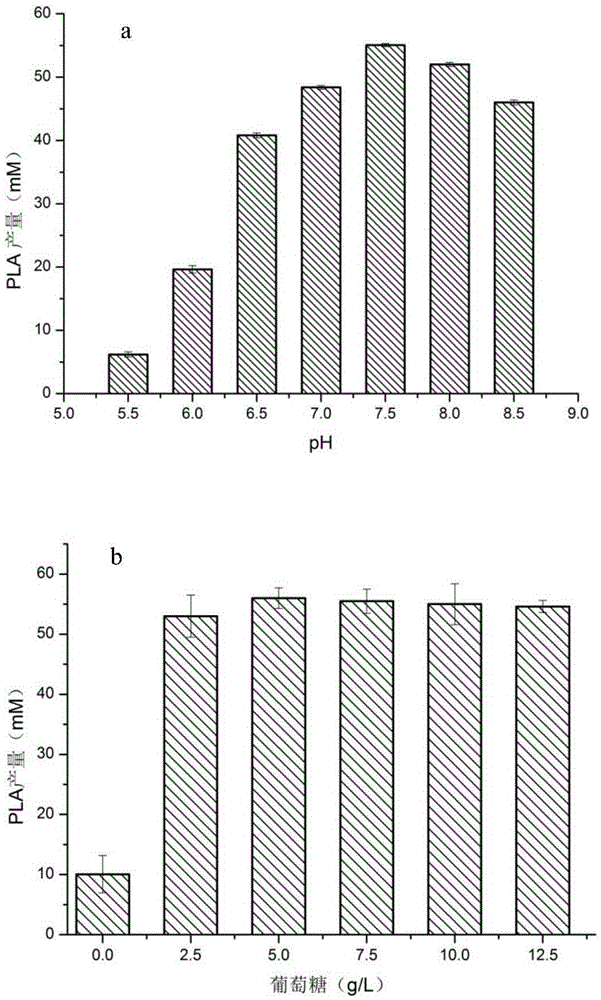
[0078] Example 3. Optimization of the whole-cell transformation reaction of D-lactate dehydrogenase mutant M307L
[0079] Cultivate the recombinant strain JM109(DE3) / M307L to the exponential phase, transfer it to fresh LB liquid medium with 1% inoculum, and the medium contains kanamycin with a concentration of 40mg / L, and cultivate until the OD600nm reaches about 0.6, and then add the final IPTG at a concentration of 1 mM was incubated at 16 °C for 16 h. 4 ℃, 5000r / min centrifugation for 15min to collect the bacteria, wash the bacteria three times with pH7.0, 50mM phosphate buffer, and resuspend in different pH (5.5 ~ 8.5) phosphate buffer (recipe: weigh 79gNaCl, 2gKCl ,2.4gKH 2 PO 4 and 1.8gK 2 HPO 4 , dissolve in 800ml of distilled water, adjust the pH of the solution to 5.5-8.5 with HCl, and finally add distilled water to dilute to 1L. ) in, obtain pH value 5.5 bacterial suspension, pH 6 bacterial suspension, pH 6.5 bacterial suspension, pH 7 bacterial suspension, pH 7...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com